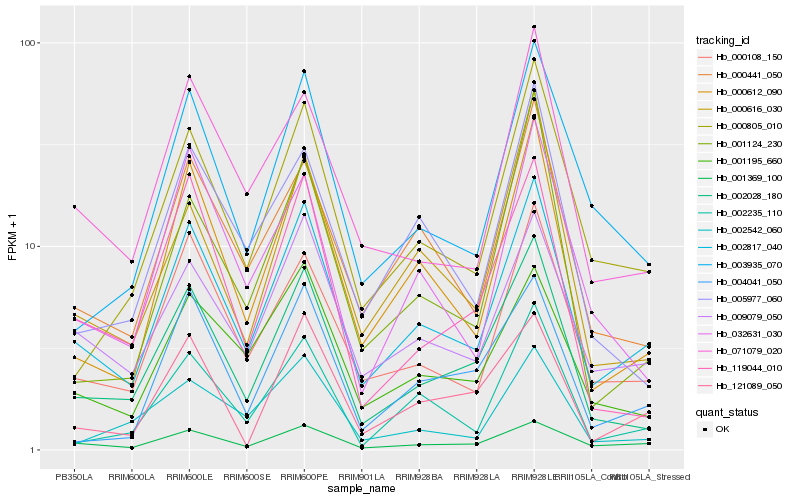
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002028_180 |
0.0 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 2 |
Hb_119044_010 |
0.0943182351 |
- |
- |
PREDICTED: stomatal closure-related actin-binding protein 2-like [Jatropha curcas] |
| 3 |
Hb_002817_040 |
0.1098053493 |
- |
- |
PREDICTED: kinesin-II 85 kDa subunit isoform X1 [Jatropha curcas] |
| 4 |
Hb_000612_090 |
0.1268253405 |
- |
- |
PREDICTED: mitochondrial carnitine/acylcarnitine carrier-like protein [Jatropha curcas] |
| 5 |
Hb_000805_010 |
0.136106114 |
- |
- |
PREDICTED: uncharacterized protein LOC105629931 [Jatropha curcas] |
| 6 |
Hb_003935_070 |
0.1388037719 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000616_030 |
0.1396547882 |
- |
- |
PREDICTED: dentin sialophosphoprotein [Jatropha curcas] |
| 8 |
Hb_000108_150 |
0.1396606809 |
- |
- |
alpha/beta hydrolase, putative [Ricinus communis] |
| 9 |
Hb_000441_050 |
0.1409428154 |
- |
- |
PREDICTED: 187-kDa microtubule-associated protein AIR9 [Jatropha curcas] |
| 10 |
Hb_001195_660 |
0.1423904257 |
- |
- |
metalloendopeptidase, putative [Ricinus communis] |
| 11 |
Hb_005977_060 |
0.1426170265 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_071079_020 |
0.1464519391 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 13 |
Hb_032631_030 |
0.1484958138 |
- |
- |
phosphatidic acid phosphatase, putative [Ricinus communis] |
| 14 |
Hb_004041_050 |
0.1490753675 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like [Jatropha curcas] |
| 15 |
Hb_009079_050 |
0.1498854352 |
- |
- |
PREDICTED: V-type proton ATPase subunit a3-like [Jatropha curcas] |
| 16 |
Hb_002542_060 |
0.1536151897 |
- |
- |
PREDICTED: wee1-like protein kinase [Jatropha curcas] |
| 17 |
Hb_001124_230 |
0.154683115 |
- |
- |
PREDICTED: divinyl chlorophyllide a 8-vinyl-reductase, chloroplastic [Jatropha curcas] |
| 18 |
Hb_121089_050 |
0.1552620767 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002235_110 |
0.1554931747 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 20 |
Hb_001369_100 |
0.1556910604 |
- |
- |
hypothetical protein JCGZ_26280 [Jatropha curcas] |