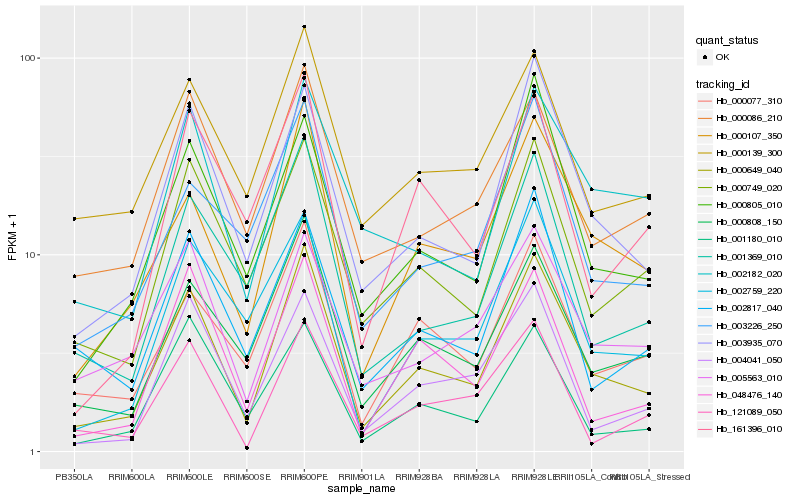
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000649_040 |
0.0 |
- |
- |
PREDICTED: dynamin-related protein 1C [Jatropha curcas] |
| 2 |
Hb_000107_350 |
0.1024740861 |
- |
- |
PREDICTED: uncharacterized protein LOC105635576 [Jatropha curcas] |
| 3 |
Hb_003935_070 |
0.1045571068 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000808_150 |
0.1180488055 |
- |
- |
PREDICTED: uncharacterized protein LOC105647444 [Jatropha curcas] |
| 5 |
Hb_000077_310 |
0.1249400529 |
- |
- |
PREDICTED: clathrin light chain 1 [Jatropha curcas] |
| 6 |
Hb_000749_020 |
0.1268164341 |
- |
- |
Nucleotide-diphospho-sugar transferases superfamily protein isoform 1 [Theobroma cacao] |
| 7 |
Hb_000805_010 |
0.1276280072 |
- |
- |
PREDICTED: uncharacterized protein LOC105629931 [Jatropha curcas] |
| 8 |
Hb_001369_010 |
0.1312292173 |
- |
- |
Nodulation receptor kinase precursor, putative [Ricinus communis] |
| 9 |
Hb_002759_220 |
0.133636973 |
- |
- |
altered response to gravity (arg1), plant, putative [Ricinus communis] |
| 10 |
Hb_048476_140 |
0.1349087005 |
- |
- |
PREDICTED: dynamin-related protein 1C [Jatropha curcas] |
| 11 |
Hb_004041_050 |
0.1390011067 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like [Jatropha curcas] |
| 12 |
Hb_000086_210 |
0.139428711 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 13 |
Hb_000139_300 |
0.1414048045 |
- |
- |
hypothetical protein POPTR_0008s19950g [Populus trichocarpa] |
| 14 |
Hb_002817_040 |
0.1415367106 |
- |
- |
PREDICTED: kinesin-II 85 kDa subunit isoform X1 [Jatropha curcas] |
| 15 |
Hb_005563_010 |
0.1415438953 |
- |
- |
PREDICTED: kinesin-II 85 kDa subunit isoform X1 [Jatropha curcas] |
| 16 |
Hb_003226_250 |
0.141715318 |
- |
- |
PREDICTED: UDP-glucuronic acid decarboxylase 2 [Jatropha curcas] |
| 17 |
Hb_002182_020 |
0.1453981747 |
- |
- |
Thylakoid lumenal 17.4 kDa protein, chloroplast precursor, putative [Ricinus communis] |
| 18 |
Hb_121089_050 |
0.1462902137 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001180_010 |
0.1478750051 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 9 [Jatropha curcas] |
| 20 |
Hb_161396_010 |
0.1485446279 |
- |
- |
hypothetical protein F383_26394 [Gossypium arboreum] |