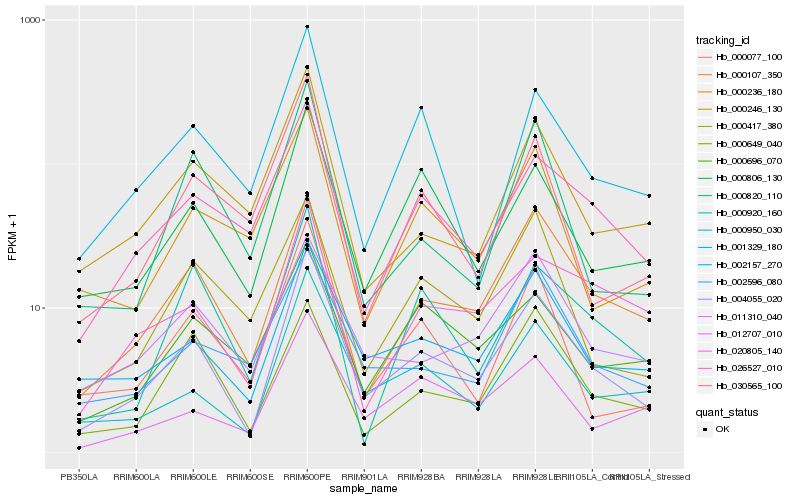
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004055_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105631433 [Jatropha curcas] |
| 2 |
Hb_000950_030 |
0.1275555277 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g10290 [Nelumbo nucifera] |
| 3 |
Hb_002157_270 |
0.1336130669 |
- |
- |
PREDICTED: transmembrane protein 87B [Jatropha curcas] |
| 4 |
Hb_000107_350 |
0.1419237209 |
- |
- |
PREDICTED: uncharacterized protein LOC105635576 [Jatropha curcas] |
| 5 |
Hb_000806_130 |
0.1504423118 |
- |
- |
Adenosine kinase 2 [Theobroma cacao] |
| 6 |
Hb_026527_010 |
0.1507587724 |
- |
- |
PREDICTED: caffeoylshikimate esterase-like [Populus euphratica] |
| 7 |
Hb_000077_100 |
0.1549768638 |
- |
- |
PREDICTED: mannan endo-1,4-beta-mannosidase 2-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_011310_040 |
0.1642455845 |
transcription factor |
TF Family: bZIP |
transcription factor hy5, putative [Ricinus communis] |
| 9 |
Hb_012707_010 |
0.1646254381 |
- |
- |
PREDICTED: uncharacterized protein C594.04c [Jatropha curcas] |
| 10 |
Hb_000246_130 |
0.1651958855 |
- |
- |
tubulin alpha chain, putative [Ricinus communis] |
| 11 |
Hb_001329_180 |
0.1653465605 |
- |
- |
S-adenosylmethionine synthetase 1 family protein [Populus trichocarpa] |
| 12 |
Hb_020805_140 |
0.168918204 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At5g42360-like [Jatropha curcas] |
| 13 |
Hb_002596_080 |
0.1734889461 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At5g04160 [Jatropha curcas] |
| 14 |
Hb_000417_380 |
0.1735027217 |
- |
- |
SecY protein transport family protein [Theobroma cacao] |
| 15 |
Hb_000649_040 |
0.1736067626 |
- |
- |
PREDICTED: dynamin-related protein 1C [Jatropha curcas] |
| 16 |
Hb_000696_070 |
0.1742531271 |
- |
- |
PREDICTED: aspartic proteinase-like protein 1 [Jatropha curcas] |
| 17 |
Hb_000920_160 |
0.1744092101 |
- |
- |
annexin [Manihot esculenta] |
| 18 |
Hb_000820_110 |
0.1744539697 |
- |
- |
UDP-XYL synthase 6 isoform 1 [Theobroma cacao] |
| 19 |
Hb_030565_100 |
0.17463771 |
- |
- |
serine hydroxymethyltransferase, putative [Ricinus communis] |
| 20 |
Hb_000236_180 |
0.1751206673 |
- |
- |
serine hydroxymethyltransferase, putative [Ricinus communis] |