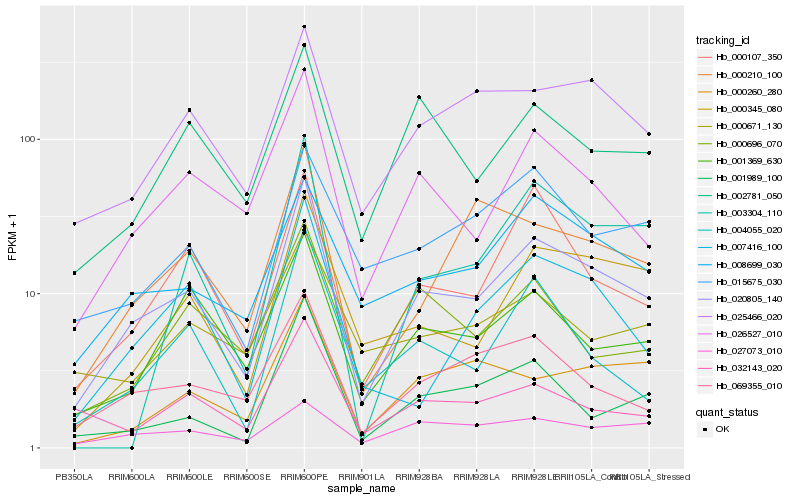
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_020805_140 |
0.0 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At5g42360-like [Jatropha curcas] |
| 2 |
Hb_026527_010 |
0.1368563841 |
- |
- |
PREDICTED: caffeoylshikimate esterase-like [Populus euphratica] |
| 3 |
Hb_000107_350 |
0.1515546686 |
- |
- |
PREDICTED: uncharacterized protein LOC105635576 [Jatropha curcas] |
| 4 |
Hb_069355_010 |
0.154286457 |
- |
- |
NBS-LRR resistance protein RGH2 [Manihot esculenta] |
| 5 |
Hb_025466_020 |
0.1546336738 |
- |
- |
tubulin beta-6 [Brassica oleracea var. italica] |
| 6 |
Hb_000345_080 |
0.1548559488 |
- |
- |
PREDICTED: protein RALF-like 33 [Jatropha curcas] |
| 7 |
Hb_008699_030 |
0.1594629125 |
- |
- |
hypothetical protein CISIN_1g017963mg [Citrus sinensis] |
| 8 |
Hb_001369_630 |
0.1632992089 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 3-like [Jatropha curcas] |
| 9 |
Hb_002781_050 |
0.1676783137 |
- |
- |
cytochrome B5 isoform 1, putative [Ricinus communis] |
| 10 |
Hb_007416_100 |
0.168219923 |
- |
- |
PREDICTED: probable xyloglucan glycosyltransferase 5 [Jatropha curcas] |
| 11 |
Hb_004055_020 |
0.168918204 |
- |
- |
PREDICTED: uncharacterized protein LOC105631433 [Jatropha curcas] |
| 12 |
Hb_001989_100 |
0.1698959623 |
- |
- |
PREDICTED: uncharacterized protein LOC105636214 [Jatropha curcas] |
| 13 |
Hb_000210_100 |
0.1717738215 |
- |
- |
pepsin A, putative [Ricinus communis] |
| 14 |
Hb_000260_280 |
0.1754560376 |
- |
- |
auxin:hydrogen symporter, putative [Ricinus communis] |
| 15 |
Hb_015675_030 |
0.1797832077 |
- |
- |
PREDICTED: UMP-CMP kinase 3 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000696_070 |
0.1802585268 |
- |
- |
PREDICTED: aspartic proteinase-like protein 1 [Jatropha curcas] |
| 17 |
Hb_032143_020 |
0.1806495115 |
- |
- |
hypothetical protein POPTR_0009s06310g [Populus trichocarpa] |
| 18 |
Hb_000671_130 |
0.1837393984 |
- |
- |
PREDICTED: CMP-sialic acid transporter 2 isoform X2 [Jatropha curcas] |
| 19 |
Hb_003304_110 |
0.1850949716 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_027073_010 |
0.1877457202 |
- |
- |
PREDICTED: protein CHROMOSOME TRANSMISSION FIDELITY 7 [Jatropha curcas] |