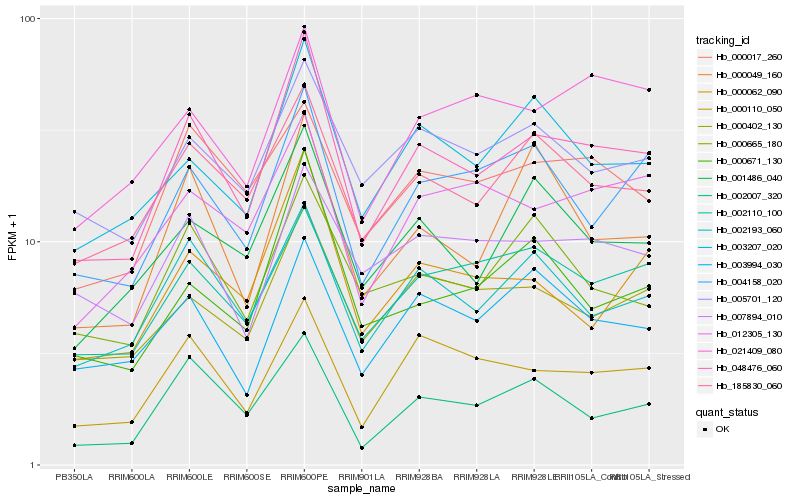
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_048476_060 |
0.0 |
- |
- |
PREDICTED: CRAL-TRIO domain-containing protein YKL091C-like isoform X2 [Jatropha curcas] |
| 2 |
Hb_002193_060 |
0.1042699032 |
- |
- |
PREDICTED: uncharacterized protein C4orf29 homolog [Pyrus x bretschneideri] |
| 3 |
Hb_185830_060 |
0.1045562209 |
- |
- |
PREDICTED: ras-related protein RABB1c [Fragaria vesca subsp. vesca] |
| 4 |
Hb_003207_020 |
0.1085572089 |
- |
- |
PREDICTED: bifunctional dTDP-4-dehydrorhamnose 3,5-epimerase/dTDP-4-dehydrorhamnose reductase [Jatropha curcas] |
| 5 |
Hb_000110_050 |
0.1089152987 |
- |
- |
- |
| 6 |
Hb_000402_130 |
0.1092161366 |
- |
- |
PREDICTED: AP-1 complex subunit mu-2 [Jatropha curcas] |
| 7 |
Hb_002007_320 |
0.1110720227 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase ANP1 [Jatropha curcas] |
| 8 |
Hb_001486_040 |
0.112068328 |
- |
- |
PREDICTED: uncharacterized protein LOC105632624 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000062_090 |
0.1129908107 |
- |
- |
PREDICTED: uncharacterized protein LOC105644743 isoform X2 [Jatropha curcas] |
| 10 |
Hb_004158_020 |
0.1159582296 |
- |
- |
Rab1 [Hevea brasiliensis] |
| 11 |
Hb_005701_120 |
0.1187376095 |
- |
- |
PREDICTED: V-type proton ATPase 16 kDa proteolipid subunit [Cucumis sativus] |
| 12 |
Hb_000671_130 |
0.1198116388 |
- |
- |
PREDICTED: CMP-sialic acid transporter 2 isoform X2 [Jatropha curcas] |
| 13 |
Hb_012305_130 |
0.1218674773 |
- |
- |
PREDICTED: GTP-binding protein SAR1A [Jatropha curcas] |
| 14 |
Hb_002110_100 |
0.1223531434 |
- |
- |
PREDICTED: pyridoxine/pyridoxamine 5'-phosphate oxidase 1, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000017_260 |
0.1242383107 |
- |
- |
hypothetical protein POPTR_0003s13070g [Populus trichocarpa] |
| 16 |
Hb_003994_030 |
0.1245407365 |
- |
- |
PREDICTED: organic cation/carnitine transporter 7 [Jatropha curcas] |
| 17 |
Hb_007894_010 |
0.1256190988 |
- |
- |
PREDICTED: uncharacterized protein LOC105632963 [Jatropha curcas] |
| 18 |
Hb_000665_180 |
0.1266691123 |
- |
- |
PREDICTED: ER membrane protein complex subunit 7 homolog isoform X2 [Jatropha curcas] |
| 19 |
Hb_000049_160 |
0.1276973494 |
- |
- |
tyrosine decarboxylase family protein [Populus trichocarpa] |
| 20 |
Hb_021409_080 |
0.129817036 |
- |
- |
PREDICTED: ORM1-like protein 1 [Jatropha curcas] |