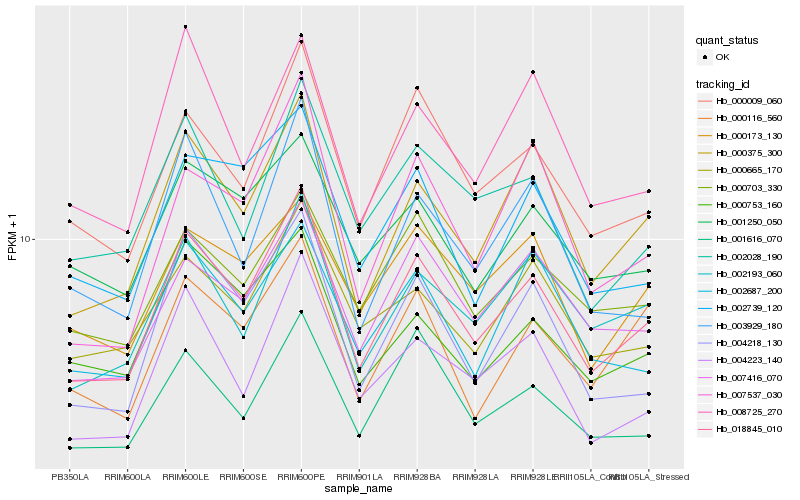
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_018845_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642055 [Jatropha curcas] |
| 2 |
Hb_000375_300 |
0.0705198058 |
- |
- |
PREDICTED: cyanate hydratase [Jatropha curcas] |
| 3 |
Hb_000009_060 |
0.0757034684 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 1B [Jatropha curcas] |
| 4 |
Hb_002193_060 |
0.0858814035 |
- |
- |
PREDICTED: uncharacterized protein C4orf29 homolog [Pyrus x bretschneideri] |
| 5 |
Hb_004218_130 |
0.0865018402 |
- |
- |
PREDICTED: uridine 5'-monophosphate synthase [Jatropha curcas] |
| 6 |
Hb_007416_070 |
0.0878104486 |
transcription factor |
TF Family: B3 |
hypothetical protein JCGZ_07038 [Jatropha curcas] |
| 7 |
Hb_001616_070 |
0.091206236 |
- |
- |
PREDICTED: uncharacterized protein LOC105644365 [Jatropha curcas] |
| 8 |
Hb_004223_140 |
0.0920497551 |
- |
- |
endo-1,4-beta-glucanase, putative [Ricinus communis] |
| 9 |
Hb_002028_190 |
0.0949153796 |
- |
- |
PREDICTED: 26S protease regulatory subunit 7-like [Jatropha curcas] |
| 10 |
Hb_002739_120 |
0.0952564921 |
- |
- |
PREDICTED: zinc finger protein ZPR1-like [Jatropha curcas] |
| 11 |
Hb_000116_560 |
0.0956075647 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 [Jatropha curcas] |
| 12 |
Hb_007537_030 |
0.096214617 |
- |
- |
PREDICTED: gamma aminobutyrate transaminase 3, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000753_160 |
0.0965843044 |
- |
- |
PREDICTED: uncharacterized protein LOC105640669 [Jatropha curcas] |
| 14 |
Hb_000665_170 |
0.0967246201 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 10 [Jatropha curcas] |
| 15 |
Hb_000703_330 |
0.0975607381 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_003929_180 |
0.0989222527 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g17430 isoform X1 [Glycine max] |
| 17 |
Hb_002687_200 |
0.0992677663 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001250_050 |
0.1001188485 |
- |
- |
PREDICTED: uncharacterized protein LOC105644623 isoform X1 [Jatropha curcas] |
| 19 |
Hb_008725_270 |
0.1009643767 |
- |
- |
PREDICTED: probable protein disulfide-isomerase A6 [Jatropha curcas] |
| 20 |
Hb_000173_130 |
0.1025144816 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase LUL4 isoform X1 [Jatropha curcas] |