| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
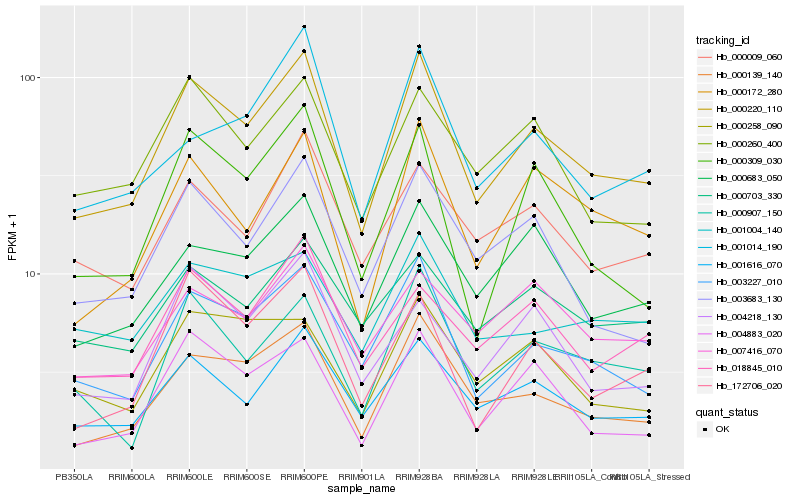
Hb_000220_110 |
0.0 |
- |
- |
fructose-bisphosphate aldolase, putative [Ricinus communis] |
| 2 |
Hb_001616_070 |
0.0849925592 |
- |
- |
PREDICTED: uncharacterized protein LOC105644365 [Jatropha curcas] |
| 3 |
Hb_003227_010 |
0.0859239673 |
- |
- |
PREDICTED: dynamin-related protein 1E-like [Jatropha curcas] |
| 4 |
Hb_000139_140 |
0.0910716298 |
- |
- |
Acyl-CoA synthetase [Ricinus communis] |
| 5 |
Hb_000172_280 |
0.1007398073 |
- |
- |
aspartate aminotransferase, putative [Ricinus communis] |
| 6 |
Hb_000683_050 |
0.108514387 |
- |
- |
PREDICTED: probable carbohydrate esterase At4g34215 [Jatropha curcas] |
| 7 |
Hb_003683_130 |
0.1104841069 |
- |
- |
PREDICTED: enolase 1, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000260_400 |
0.1138465633 |
- |
- |
PREDICTED: 2,3-bisphosphoglycerate-independent phosphoglycerate mutase [Jatropha curcas] |
| 9 |
Hb_004883_020 |
0.1139879812 |
- |
- |
PREDICTED: uncharacterized protein LOC105647351 [Jatropha curcas] |
| 10 |
Hb_000309_030 |
0.1142887172 |
- |
- |
PREDICTED: formate dehydrogenase, mitochondrial [Jatropha curcas] |
| 11 |
Hb_000009_060 |
0.1143661326 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 1B [Jatropha curcas] |
| 12 |
Hb_018845_010 |
0.1176495669 |
- |
- |
PREDICTED: uncharacterized protein LOC105642055 [Jatropha curcas] |
| 13 |
Hb_000907_150 |
0.1177813012 |
- |
- |
hypothetical protein L484_004295 [Morus notabilis] |
| 14 |
Hb_172706_020 |
0.1205510714 |
- |
- |
PREDICTED: protein ABIL2 [Jatropha curcas] |
| 15 |
Hb_004218_130 |
0.124858063 |
- |
- |
PREDICTED: uridine 5'-monophosphate synthase [Jatropha curcas] |
| 16 |
Hb_007416_070 |
0.1255701633 |
transcription factor |
TF Family: B3 |
hypothetical protein JCGZ_07038 [Jatropha curcas] |
| 17 |
Hb_000258_090 |
0.1258367343 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X2 [Jatropha curcas] |
| 18 |
Hb_001004_140 |
0.1262808979 |
- |
- |
hypothetical protein JCGZ_26893 [Jatropha curcas] |
| 19 |
Hb_000703_330 |
0.127019902 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001014_190 |
0.1275826405 |
- |
- |
PREDICTED: uncharacterized protein LOC105647294 [Jatropha curcas] |