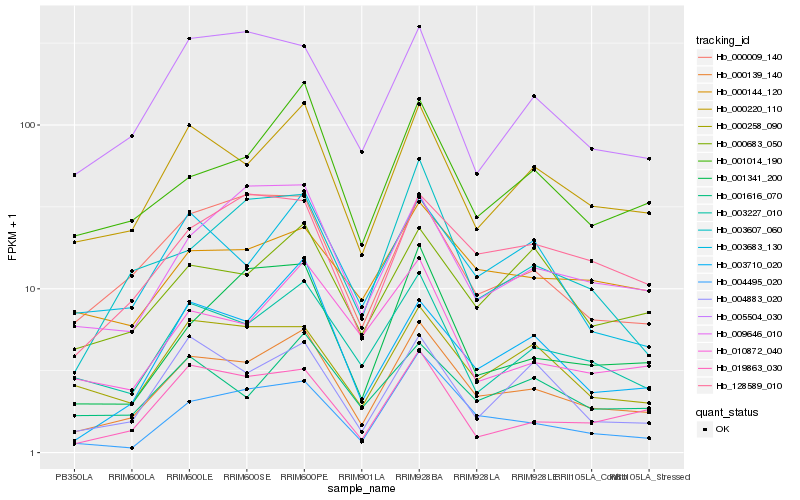
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000139_140 |
0.0 |
- |
- |
Acyl-CoA synthetase [Ricinus communis] |
| 2 |
Hb_000220_110 |
0.0910716298 |
- |
- |
fructose-bisphosphate aldolase, putative [Ricinus communis] |
| 3 |
Hb_003227_010 |
0.1136057889 |
- |
- |
PREDICTED: dynamin-related protein 1E-like [Jatropha curcas] |
| 4 |
Hb_001014_190 |
0.1255194899 |
- |
- |
PREDICTED: uncharacterized protein LOC105647294 [Jatropha curcas] |
| 5 |
Hb_004495_020 |
0.1273212547 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 6 |
Hb_000009_140 |
0.1294492353 |
transcription factor |
TF Family: BES1 |
BRASSINAZOLE-RESISTANT 1 protein, putative [Ricinus communis] |
| 7 |
Hb_009646_010 |
0.1329804626 |
- |
- |
PREDICTED: hyoscyamine 6-dioxygenase-like [Jatropha curcas] |
| 8 |
Hb_128589_010 |
0.133062803 |
- |
- |
PREDICTED: cyclin-dependent kinase inhibitor 5 [Jatropha curcas] |
| 9 |
Hb_003683_130 |
0.133780347 |
- |
- |
PREDICTED: enolase 1, chloroplastic [Jatropha curcas] |
| 10 |
Hb_001616_070 |
0.1359946364 |
- |
- |
PREDICTED: uncharacterized protein LOC105644365 [Jatropha curcas] |
| 11 |
Hb_003710_020 |
0.1363982051 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g53440 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000683_050 |
0.1367025558 |
- |
- |
PREDICTED: probable carbohydrate esterase At4g34215 [Jatropha curcas] |
| 13 |
Hb_000258_090 |
0.1368423943 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X2 [Jatropha curcas] |
| 14 |
Hb_001341_200 |
0.1383991987 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 4 [Jatropha curcas] |
| 15 |
Hb_019863_030 |
0.1411781445 |
- |
- |
hypothetical protein JCGZ_00393 [Jatropha curcas] |
| 16 |
Hb_004883_020 |
0.1417163339 |
- |
- |
PREDICTED: uncharacterized protein LOC105647351 [Jatropha curcas] |
| 17 |
Hb_003607_060 |
0.1427894274 |
- |
- |
PREDICTED: alcohol dehydrogenase-like [Jatropha curcas] |
| 18 |
Hb_010872_040 |
0.1430860107 |
- |
- |
hypothetical protein F775_31993 [Aegilops tauschii] |
| 19 |
Hb_005504_030 |
0.1440051798 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 2-like [Pyrus x bretschneideri] |
| 20 |
Hb_000144_120 |
0.1459274558 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |