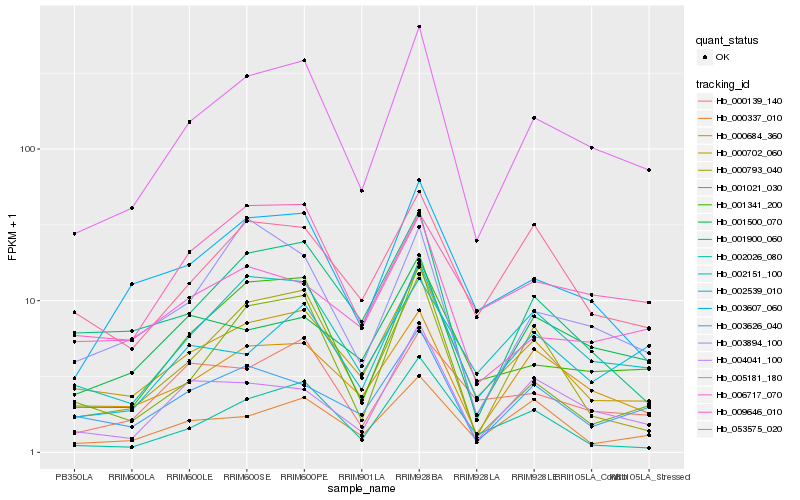
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005181_180 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001341_200 |
0.1113877287 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 4 [Jatropha curcas] |
| 3 |
Hb_000702_060 |
0.1191439788 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000684_360 |
0.1305532617 |
transcription factor |
TF Family: AP2 |
hypothetical protein JCGZ_16454 [Jatropha curcas] |
| 5 |
Hb_003607_060 |
0.1323161807 |
- |
- |
PREDICTED: alcohol dehydrogenase-like [Jatropha curcas] |
| 6 |
Hb_000793_040 |
0.1472234647 |
- |
- |
PREDICTED: transcription factor LHW [Jatropha curcas] |
| 7 |
Hb_001900_060 |
0.1497506396 |
- |
- |
PREDICTED: neutral alpha-glucosidase C [Jatropha curcas] |
| 8 |
Hb_002539_010 |
0.1511606456 |
- |
- |
hypothetical protein JCGZ_15711 [Jatropha curcas] |
| 9 |
Hb_002151_100 |
0.1521195698 |
- |
- |
PREDICTED: uncharacterized protein LOC105637951 [Jatropha curcas] |
| 10 |
Hb_004041_100 |
0.1546582784 |
- |
- |
- |
| 11 |
Hb_003626_040 |
0.1550126978 |
- |
- |
PREDICTED: probable 2-oxoglutarate-dependent dioxygenase At3g49630 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000139_140 |
0.1579396295 |
- |
- |
Acyl-CoA synthetase [Ricinus communis] |
| 13 |
Hb_003894_100 |
0.1586547505 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 1-like [Jatropha curcas] |
| 14 |
Hb_001021_030 |
0.1602561119 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 15 |
Hb_053575_020 |
0.1605475873 |
- |
- |
PREDICTED: RNA-binding protein 39 [Jatropha curcas] |
| 16 |
Hb_000337_010 |
0.1609038713 |
- |
- |
PREDICTED: proteasome activator subunit 4 isoform X2 [Jatropha curcas] |
| 17 |
Hb_006717_070 |
0.1610522731 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor CPC-like [Jatropha curcas] |
| 18 |
Hb_009646_010 |
0.1622613713 |
- |
- |
PREDICTED: hyoscyamine 6-dioxygenase-like [Jatropha curcas] |
| 19 |
Hb_001500_070 |
0.1631593359 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_002026_080 |
0.1634307234 |
desease resistance |
Gene Name: TIR_2 |
nucleoside-triphosphatase, putative [Ricinus communis] |