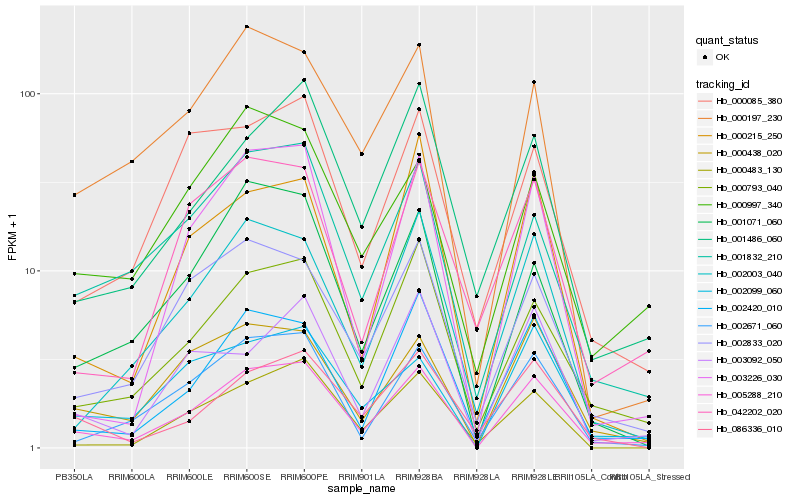
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005288_210 |
0.0 |
- |
- |
hypothetical protein VITISV_017217 [Vitis vinifera] |
| 2 |
Hb_000793_040 |
0.1193891958 |
- |
- |
PREDICTED: transcription factor LHW [Jatropha curcas] |
| 3 |
Hb_000438_020 |
0.1306797427 |
- |
- |
leucine-rich repeat-containing protein 2, lrrc2, putative [Ricinus communis] |
| 4 |
Hb_002420_010 |
0.1309314903 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 5 |
Hb_002671_060 |
0.1346808935 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD3-1 [Jatropha curcas] |
| 6 |
Hb_003226_030 |
0.1357141016 |
- |
- |
sucrose synthase 5 [Hevea brasiliensis] |
| 7 |
Hb_002833_020 |
0.1394662079 |
desease resistance |
Gene Name: LRR_4 |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 8 |
Hb_002003_040 |
0.1410917393 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 isoform X1 [Jatropha curcas] |
| 9 |
Hb_002099_060 |
0.1424331803 |
- |
- |
Protein AFR, putative [Ricinus communis] |
| 10 |
Hb_001832_210 |
0.1434413138 |
- |
- |
PREDICTED: venom phosphodiesterase 2 [Jatropha curcas] |
| 11 |
Hb_042202_020 |
0.1483851695 |
- |
- |
Eukaryotic release factor 1-3 isoform 1 [Theobroma cacao] |
| 12 |
Hb_001071_060 |
0.1517154267 |
- |
- |
Trehalose phosphatase/synthase 5 isoform 1 [Theobroma cacao] |
| 13 |
Hb_000483_130 |
0.152997152 |
- |
- |
PREDICTED: uncharacterized protein LOC105644062 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001486_060 |
0.1533283188 |
- |
- |
PREDICTED: uncharacterized protein LOC105632622 [Jatropha curcas] |
| 15 |
Hb_003092_050 |
0.1546933019 |
- |
- |
PREDICTED: putative chloride channel-like protein CLC-g [Jatropha curcas] |
| 16 |
Hb_000197_230 |
0.1550196191 |
transcription factor |
TF Family: bZIP |
Ocs element-binding factor, putative [Ricinus communis] |
| 17 |
Hb_000215_250 |
0.1550650492 |
- |
- |
Acid phosphatase 1 precursor, putative [Ricinus communis] |
| 18 |
Hb_000085_380 |
0.1566834637 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_086336_010 |
0.1571533564 |
- |
- |
hypothetical protein CISIN_1g022501mg [Citrus sinensis] |
| 20 |
Hb_000997_340 |
0.1574352903 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |