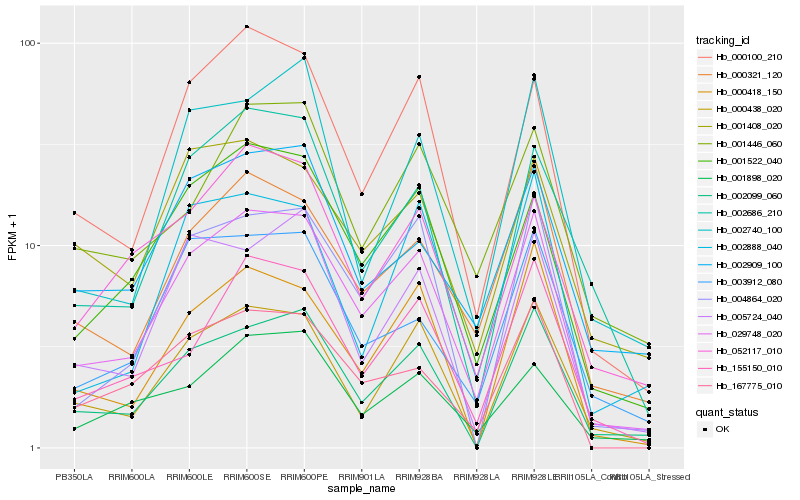
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_029748_020 |
0.0 |
- |
- |
PREDICTED: ent-kaur-16-ene synthase, chloroplastic isoform X1 [Jatropha curcas] |
| 2 |
Hb_000100_210 |
0.0971551398 |
- |
- |
PREDICTED: selenium-binding protein 1-like [Jatropha curcas] |
| 3 |
Hb_000321_120 |
0.0988274736 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g59700 [Jatropha curcas] |
| 4 |
Hb_001522_040 |
0.0988615792 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 5 |
Hb_002099_060 |
0.1032284088 |
- |
- |
Protein AFR, putative [Ricinus communis] |
| 6 |
Hb_000438_020 |
0.11256622 |
- |
- |
leucine-rich repeat-containing protein 2, lrrc2, putative [Ricinus communis] |
| 7 |
Hb_155150_010 |
0.1140610759 |
- |
- |
kinase, putative [Ricinus communis] |
| 8 |
Hb_004864_020 |
0.1148430099 |
- |
- |
Polyprotein, putative [Solanum demissum] |
| 9 |
Hb_167775_010 |
0.1171882284 |
- |
- |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 10 |
Hb_000418_150 |
0.1244197431 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 11 |
Hb_005724_040 |
0.1260919142 |
- |
- |
hypothetical protein JCGZ_13142 [Jatropha curcas] |
| 12 |
Hb_002686_210 |
0.1272307219 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 13 |
Hb_001898_020 |
0.1273517375 |
- |
- |
PREDICTED: uncharacterized protein LOC105629659 [Jatropha curcas] |
| 14 |
Hb_003912_080 |
0.1287508532 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 15 |
Hb_052117_010 |
0.1290080661 |
- |
- |
kinase, putative [Ricinus communis] |
| 16 |
Hb_002909_100 |
0.1324883448 |
- |
- |
PREDICTED: uncharacterized protein LOC105648153 [Jatropha curcas] |
| 17 |
Hb_002740_100 |
0.1333991293 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 18 |
Hb_002888_040 |
0.13393095 |
- |
- |
hypothetical protein B456_009G267200 [Gossypium raimondii] |
| 19 |
Hb_001446_060 |
0.1352488031 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 20 |
Hb_001408_020 |
0.1375873589 |
- |
- |
PREDICTED: phosphatidylserine decarboxylase proenzyme 2-like [Jatropha curcas] |