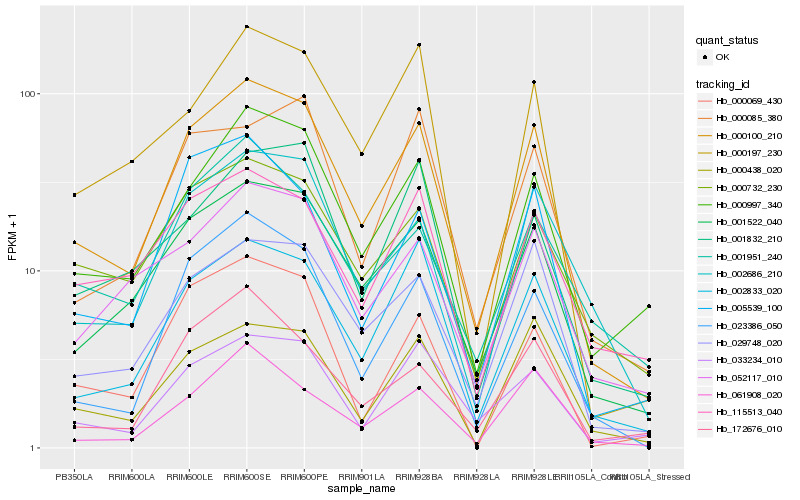
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000100_210 |
0.0 |
- |
- |
PREDICTED: selenium-binding protein 1-like [Jatropha curcas] |
| 2 |
Hb_001522_040 |
0.0879644289 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 3 |
Hb_029748_020 |
0.0971551398 |
- |
- |
PREDICTED: ent-kaur-16-ene synthase, chloroplastic isoform X1 [Jatropha curcas] |
| 4 |
Hb_002833_020 |
0.1036461397 |
desease resistance |
Gene Name: LRR_4 |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 5 |
Hb_000997_340 |
0.1043946089 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 6 |
Hb_023386_050 |
0.1106244732 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_002686_210 |
0.1143581056 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 8 |
Hb_000438_020 |
0.1231591141 |
- |
- |
leucine-rich repeat-containing protein 2, lrrc2, putative [Ricinus communis] |
| 9 |
Hb_033234_010 |
0.1237287064 |
desease resistance |
Gene Name: LRR_8 |
Disease resistance protein RPP8 [Theobroma cacao] |
| 10 |
Hb_000732_230 |
0.1249474721 |
- |
- |
PREDICTED: protein TRANSPORT INHIBITOR RESPONSE 1-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_000197_230 |
0.1269247446 |
transcription factor |
TF Family: bZIP |
Ocs element-binding factor, putative [Ricinus communis] |
| 12 |
Hb_052117_010 |
0.1278214806 |
- |
- |
kinase, putative [Ricinus communis] |
| 13 |
Hb_005539_100 |
0.1281711508 |
- |
- |
nucleoside transporter, putative [Ricinus communis] |
| 14 |
Hb_061908_020 |
0.1310526457 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 15 |
Hb_172676_010 |
0.131857559 |
- |
- |
PREDICTED: putative disease resistance protein At3g14460 [Jatropha curcas] |
| 16 |
Hb_115513_040 |
0.1336878976 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000085_380 |
0.1380815932 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001832_210 |
0.1384155004 |
- |
- |
PREDICTED: venom phosphodiesterase 2 [Jatropha curcas] |
| 19 |
Hb_000069_430 |
0.1384413842 |
- |
- |
PREDICTED: cyclic nucleotide-gated ion channel 1-like [Jatropha curcas] |
| 20 |
Hb_001951_240 |
0.1405029051 |
- |
- |
PREDICTED: protein MEI2-like 5 [Jatropha curcas] |