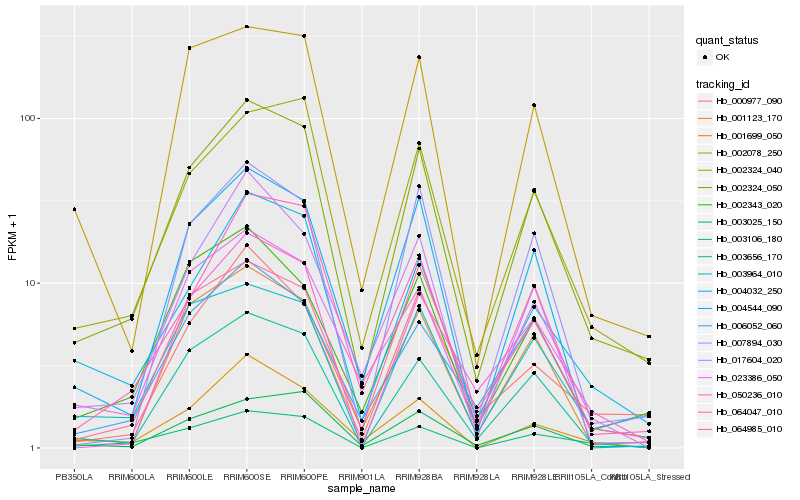
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002324_050 |
0.0 |
- |
- |
hypothetical protein AMTR_s00420p00013340 [Amborella trichopoda] |
| 2 |
Hb_003656_170 |
0.0821640553 |
- |
- |
PREDICTED: phenylalanine ammonia-lyase-like [Jatropha curcas] |
| 3 |
Hb_002324_040 |
0.1013814817 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.6 [Jatropha curcas] |
| 4 |
Hb_004032_250 |
0.1029796667 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_003964_010 |
0.1045629657 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0008s09360g [Populus trichocarpa] |
| 6 |
Hb_003025_150 |
0.109534177 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001123_170 |
0.1133008659 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_004544_090 |
0.117066347 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 9 |
Hb_023386_050 |
0.1210752826 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_064047_010 |
0.1213869993 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002078_250 |
0.1224989455 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_003106_180 |
0.124687988 |
- |
- |
Peroxidase 65 precursor, putative [Ricinus communis] |
| 13 |
Hb_006052_060 |
0.1248113471 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 14 |
Hb_001699_050 |
0.1277286814 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0005s11410g [Populus trichocarpa] |
| 15 |
Hb_002343_020 |
0.129525599 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 1 [Jatropha curcas] |
| 16 |
Hb_007894_030 |
0.1301278234 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 17 |
Hb_064985_010 |
0.1319094363 |
- |
- |
Leucine-rich repeat transmembrane protein kinase, putative [Theobroma cacao] |
| 18 |
Hb_017604_020 |
0.133558134 |
- |
- |
hypothetical protein JCGZ_22673 [Jatropha curcas] |
| 19 |
Hb_050236_010 |
0.1337922121 |
- |
- |
hypothetical protein EUGRSUZ_E02960, partial [Eucalyptus grandis] |
| 20 |
Hb_000977_090 |
0.1340460422 |
- |
- |
hypothetical protein POPTR_0010s18550g [Populus trichocarpa] |