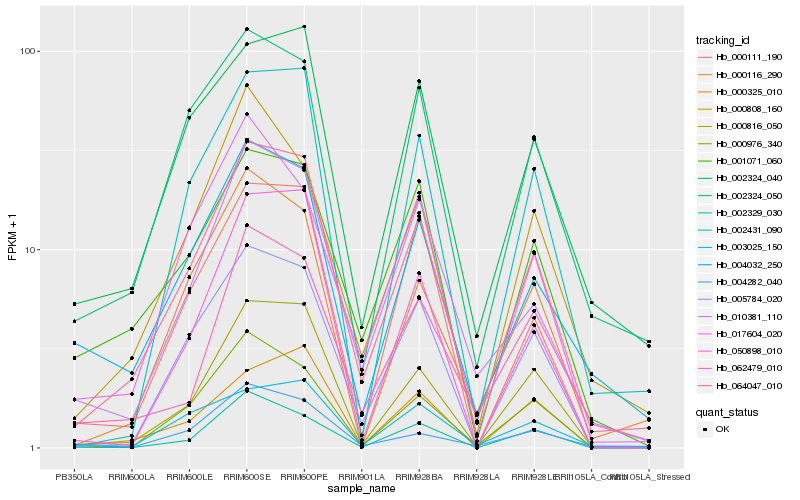
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_064047_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000816_050 |
0.0990018475 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 3 |
Hb_005784_020 |
0.1089291762 |
- |
- |
Protein MKS1, putative [Ricinus communis] |
| 4 |
Hb_002431_090 |
0.1174574744 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002324_040 |
0.1176494957 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.6 [Jatropha curcas] |
| 6 |
Hb_010381_110 |
0.1189573625 |
- |
- |
PRA1 family protein [Theobroma cacao] |
| 7 |
Hb_000976_340 |
0.1203099307 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 8 |
Hb_002324_050 |
0.1213869993 |
- |
- |
hypothetical protein AMTR_s00420p00013340 [Amborella trichopoda] |
| 9 |
Hb_004032_250 |
0.1236633815 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000116_290 |
0.1278162361 |
- |
- |
PREDICTED: proton-coupled amino acid transporter 3 [Jatropha curcas] |
| 11 |
Hb_001071_060 |
0.1303913423 |
- |
- |
Trehalose phosphatase/synthase 5 isoform 1 [Theobroma cacao] |
| 12 |
Hb_003025_150 |
0.1364455304 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000111_190 |
0.1374508208 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL1 [Jatropha curcas] |
| 14 |
Hb_004282_040 |
0.1390326846 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X2 [Jatropha curcas] |
| 15 |
Hb_000808_160 |
0.1406984236 |
- |
- |
PREDICTED: uncharacterized protein LOC105647443 [Jatropha curcas] |
| 16 |
Hb_050898_010 |
0.141894099 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |
| 17 |
Hb_002329_030 |
0.1418969438 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 2-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_000325_010 |
0.1426039475 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RGA2, putative [Ricinus communis] |
| 19 |
Hb_062479_010 |
0.1436518453 |
- |
- |
hypothetical protein PHAVU_008G071200g [Phaseolus vulgaris] |
| 20 |
Hb_017604_020 |
0.1437784013 |
- |
- |
hypothetical protein JCGZ_22673 [Jatropha curcas] |