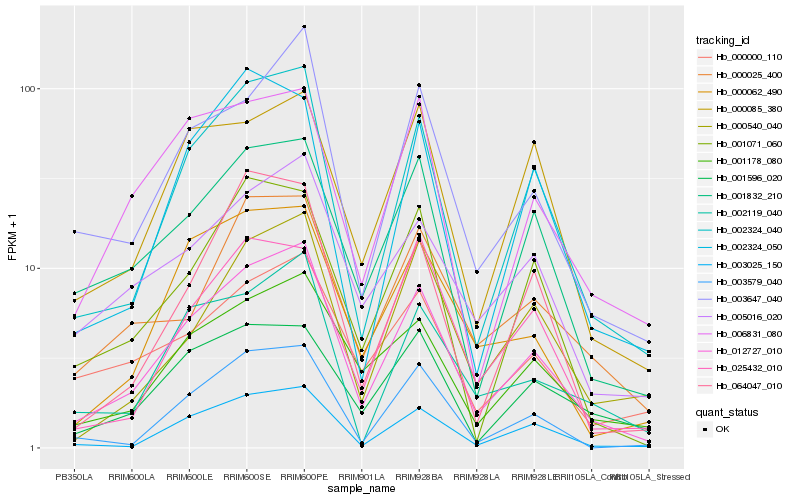
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012727_010 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 2 |
Hb_002324_040 |
0.0803141856 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.6 [Jatropha curcas] |
| 3 |
Hb_003025_150 |
0.1127888823 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_003647_040 |
0.1161618106 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001178_080 |
0.1200004928 |
- |
- |
PREDICTED: protein kinase PVPK-1 [Jatropha curcas] |
| 6 |
Hb_001832_210 |
0.1279760226 |
- |
- |
PREDICTED: venom phosphodiesterase 2 [Jatropha curcas] |
| 7 |
Hb_003579_040 |
0.1299290246 |
- |
- |
PREDICTED: monoacylglycerol lipase ABHD6 [Jatropha curcas] |
| 8 |
Hb_005016_020 |
0.132554393 |
desease resistance |
Gene Name: ABC_membrane |
ABC transporter family protein [Hevea brasiliensis] |
| 9 |
Hb_000062_490 |
0.1325911852 |
- |
- |
PREDICTED: lipoxygenase 6, chloroplastic [Jatropha curcas] |
| 10 |
Hb_002119_040 |
0.1373438432 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 11 |
Hb_001071_060 |
0.1406888877 |
- |
- |
Trehalose phosphatase/synthase 5 isoform 1 [Theobroma cacao] |
| 12 |
Hb_000540_040 |
0.1414976222 |
- |
- |
PREDICTED: chitinase 2 [Amborella trichopoda] |
| 13 |
Hb_000025_400 |
0.142780514 |
- |
- |
PREDICTED: uncharacterized protein LOC105110770 [Populus euphratica] |
| 14 |
Hb_006831_080 |
0.1438123192 |
- |
- |
PREDICTED: uncharacterized protein LOC105642126 [Jatropha curcas] |
| 15 |
Hb_000000_110 |
0.1450903349 |
- |
- |
hypothetical protein CISIN_1g0027772mg, partial [Citrus sinensis] |
| 16 |
Hb_000085_380 |
0.1488322606 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_064047_010 |
0.1518701852 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001596_020 |
0.1535833097 |
- |
- |
hypothetical protein CICLE_v10014140mg [Citrus clementina] |
| 19 |
Hb_025432_010 |
0.1542757657 |
- |
- |
hypothetical protein JCGZ_21627 [Jatropha curcas] |
| 20 |
Hb_002324_050 |
0.1544507074 |
- |
- |
hypothetical protein AMTR_s00420p00013340 [Amborella trichopoda] |