| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
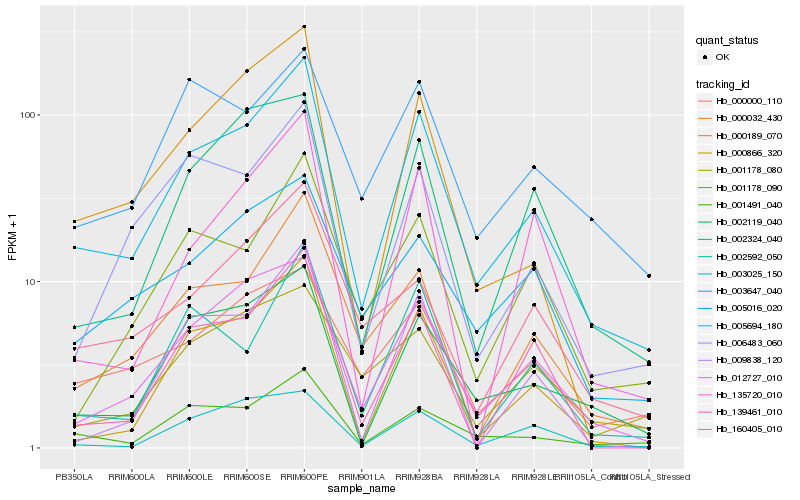
Hb_003647_040 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_012727_010 |
0.1161618106 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 3 |
Hb_001491_040 |
0.117188535 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 4 |
Hb_002119_040 |
0.1239075604 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 5 |
Hb_000032_430 |
0.1334886252 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000866_320 |
0.1358674869 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 7 |
Hb_000189_070 |
0.1457298241 |
transcription factor |
TF Family: AUX/IAA |
Auxin-induced protein 22D, putative [Ricinus communis] |
| 8 |
Hb_006483_060 |
0.1502778712 |
- |
- |
PREDICTED: protein RALF-like 34 [Jatropha curcas] |
| 9 |
Hb_002324_040 |
0.1510351565 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.6 [Jatropha curcas] |
| 10 |
Hb_000000_110 |
0.1538637982 |
- |
- |
hypothetical protein CISIN_1g0027772mg, partial [Citrus sinensis] |
| 11 |
Hb_001178_090 |
0.1546805973 |
- |
- |
Monodehydroascorbate reductase family protein [Populus trichocarpa] |
| 12 |
Hb_002592_050 |
0.1547398661 |
- |
- |
PREDICTED: LON peptidase N-terminal domain and RING finger protein 1-like [Jatropha curcas] |
| 13 |
Hb_005016_020 |
0.1559417376 |
desease resistance |
Gene Name: ABC_membrane |
ABC transporter family protein [Hevea brasiliensis] |
| 14 |
Hb_160405_010 |
0.1599699696 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 15 |
Hb_139461_010 |
0.1632184923 |
desease resistance |
Gene Name: NB-ARC |
Cc-nbs-lrr resistance protein, putative [Theobroma cacao] |
| 16 |
Hb_005694_180 |
0.166220568 |
- |
- |
PREDICTED: cytochrome b5 [Jatropha curcas] |
| 17 |
Hb_009838_120 |
0.1668458097 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA13 isoform X1 [Jatropha curcas] |
| 18 |
Hb_003025_150 |
0.1669102648 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_135720_010 |
0.1696407885 |
- |
- |
hypothetical protein JCGZ_17857 [Jatropha curcas] |
| 20 |
Hb_001178_080 |
0.1703166543 |
- |
- |
PREDICTED: protein kinase PVPK-1 [Jatropha curcas] |