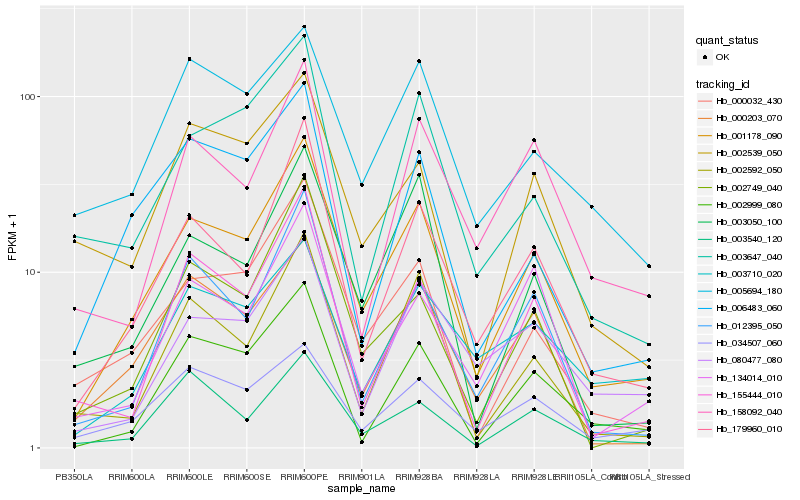
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001178_090 |
0.0 |
- |
- |
Monodehydroascorbate reductase family protein [Populus trichocarpa] |
| 2 |
Hb_179960_010 |
0.109084412 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase ATL4-like [Jatropha curcas] |
| 3 |
Hb_000032_430 |
0.1167486875 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000203_070 |
0.1309762507 |
- |
- |
PREDICTED: uncharacterized protein LOC105649674 [Jatropha curcas] |
| 5 |
Hb_012395_050 |
0.1365662254 |
- |
- |
PREDICTED: uncharacterized protein LOC105632180 [Jatropha curcas] |
| 6 |
Hb_003050_100 |
0.1390250249 |
- |
- |
PREDICTED: uncharacterized protein LOC105633987 [Jatropha curcas] |
| 7 |
Hb_002592_050 |
0.139673873 |
- |
- |
PREDICTED: LON peptidase N-terminal domain and RING finger protein 1-like [Jatropha curcas] |
| 8 |
Hb_034507_060 |
0.1414061888 |
- |
- |
PREDICTED: UPF0503 protein At3g09070, chloroplastic-like [Citrus sinensis] |
| 9 |
Hb_002999_080 |
0.1416384204 |
transcription factor |
TF Family: G2-like |
PREDICTED: accessory gland protein Acp36DE [Jatropha curcas] |
| 10 |
Hb_002749_040 |
0.1425376137 |
- |
- |
Ribosomal protein L6 family protein [Theobroma cacao] |
| 11 |
Hb_155444_010 |
0.1460058531 |
- |
- |
- |
| 12 |
Hb_006483_060 |
0.1528568175 |
- |
- |
PREDICTED: protein RALF-like 34 [Jatropha curcas] |
| 13 |
Hb_003647_040 |
0.1546805973 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_134014_010 |
0.1548377558 |
- |
- |
PREDICTED: testis-expressed sequence 10 protein isoform X2 [Jatropha curcas] |
| 15 |
Hb_005694_180 |
0.1548553925 |
- |
- |
PREDICTED: cytochrome b5 [Jatropha curcas] |
| 16 |
Hb_080477_080 |
0.1548714616 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4 isoform X1 [Gossypium raimondii] |
| 17 |
Hb_003540_120 |
0.1557502924 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 18 |
Hb_003710_020 |
0.1572652125 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g53440 isoform X1 [Jatropha curcas] |
| 19 |
Hb_158092_040 |
0.1579739562 |
- |
- |
PREDICTED: uncharacterized protein LOC105647294 [Jatropha curcas] |
| 20 |
Hb_002539_050 |
0.1583482166 |
- |
- |
PREDICTED: PLASMODESMATA CALLOSE-BINDING PROTEIN 3 [Jatropha curcas] |