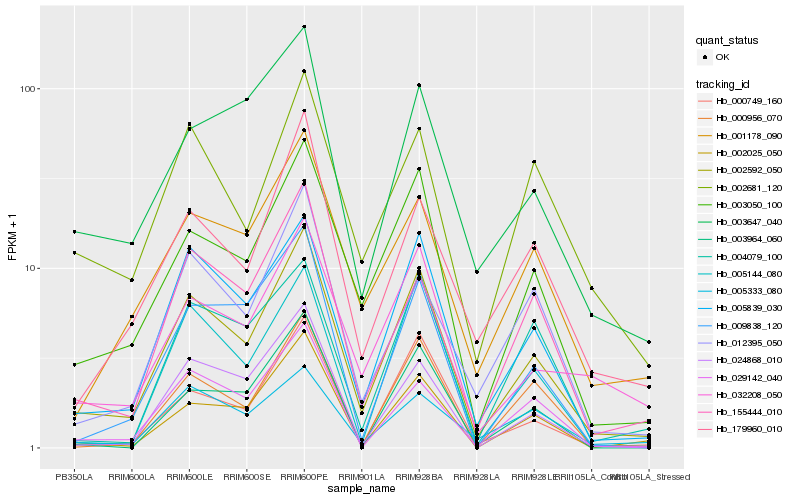
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002592_050 |
0.0 |
- |
- |
PREDICTED: LON peptidase N-terminal domain and RING finger protein 1-like [Jatropha curcas] |
| 2 |
Hb_005839_030 |
0.0960600227 |
- |
- |
PREDICTED: potassium transporter 2 [Jatropha curcas] |
| 3 |
Hb_003050_100 |
0.0966830171 |
- |
- |
PREDICTED: uncharacterized protein LOC105633987 [Jatropha curcas] |
| 4 |
Hb_032208_050 |
0.1306979559 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 4 isoform X3 [Jatropha curcas] |
| 5 |
Hb_155444_010 |
0.1356843608 |
- |
- |
- |
| 6 |
Hb_002025_050 |
0.1392101933 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001178_090 |
0.139673873 |
- |
- |
Monodehydroascorbate reductase family protein [Populus trichocarpa] |
| 8 |
Hb_000749_160 |
0.1411688942 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 9 |
Hb_029142_040 |
0.1419509293 |
- |
- |
replication factor C / DNA polymerase III gamma-tau subunit, putative [Ricinus communis] |
| 10 |
Hb_179960_010 |
0.1425118938 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase ATL4-like [Jatropha curcas] |
| 11 |
Hb_005144_080 |
0.1457542373 |
- |
- |
Serine carboxypeptidase, putative [Ricinus communis] |
| 12 |
Hb_000956_070 |
0.147165526 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g01540 [Jatropha curcas] |
| 13 |
Hb_009838_120 |
0.1480690548 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA13 isoform X1 [Jatropha curcas] |
| 14 |
Hb_024868_010 |
0.1480745034 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 15 |
Hb_002681_120 |
0.1499922176 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g14450 [Jatropha curcas] |
| 16 |
Hb_012395_050 |
0.1515063293 |
- |
- |
PREDICTED: uncharacterized protein LOC105632180 [Jatropha curcas] |
| 17 |
Hb_003647_040 |
0.1547398661 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_005333_080 |
0.1555115671 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_004079_100 |
0.1587909611 |
- |
- |
PREDICTED: uncharacterized protein LOC105637705 [Jatropha curcas] |
| 20 |
Hb_003964_060 |
0.1589200855 |
- |
- |
PREDICTED: beta-glucosidase 40 [Jatropha curcas] |