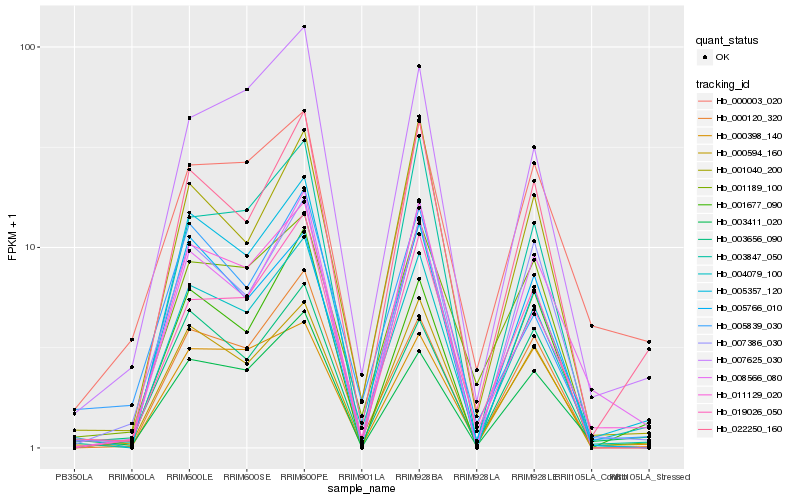
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004079_100 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105637705 [Jatropha curcas] |
| 2 |
Hb_022250_160 |
0.0769343104 |
- |
- |
respiratory burst oxidase A [Manihot esculenta] |
| 3 |
Hb_011129_020 |
0.0987369142 |
- |
- |
Beta-1,3-galactosyltransferase sqv-2, putative [Ricinus communis] |
| 4 |
Hb_003411_020 |
0.0987384039 |
- |
- |
alpha-glucosidase, putative [Ricinus communis] |
| 5 |
Hb_008566_080 |
0.1023484462 |
- |
- |
PREDICTED: uncharacterized protein LOC105647834 [Jatropha curcas] |
| 6 |
Hb_000120_320 |
0.1031256955 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 7 |
Hb_000594_160 |
0.103181381 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 8 [Jatropha curcas] |
| 8 |
Hb_001040_200 |
0.1037378991 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA1H1-like [Jatropha curcas] |
| 9 |
Hb_000398_140 |
0.1101463266 |
- |
- |
PREDICTED: methionine S-methyltransferase [Jatropha curcas] |
| 10 |
Hb_005357_120 |
0.1115414511 |
transcription factor |
TF Family: MYB-related |
- |
| 11 |
Hb_007386_030 |
0.1117833796 |
- |
- |
PREDICTED: formin-like protein 2 [Jatropha curcas] |
| 12 |
Hb_007625_030 |
0.1123308234 |
- |
- |
unknown [Medicago truncatula] |
| 13 |
Hb_001189_100 |
0.1126182342 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 2-like [Jatropha curcas] |
| 14 |
Hb_005766_010 |
0.1179274648 |
- |
- |
serine carboxypeptidase, putative [Ricinus communis] |
| 15 |
Hb_019026_050 |
0.1180189223 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000003_020 |
0.1202339709 |
- |
- |
PREDICTED: uncharacterized protein LOC105631161 [Jatropha curcas] |
| 17 |
Hb_003656_090 |
0.1205649937 |
- |
- |
PREDICTED: protein Brevis radix-like 4 [Jatropha curcas] |
| 18 |
Hb_003847_050 |
0.1206922744 |
- |
- |
PREDICTED: aquaporin SIP1-1 [Jatropha curcas] |
| 19 |
Hb_001677_090 |
0.1255076405 |
- |
- |
PREDICTED: CASP-like protein 5B2 [Jatropha curcas] |
| 20 |
Hb_005839_030 |
0.1270465699 |
- |
- |
PREDICTED: potassium transporter 2 [Jatropha curcas] |