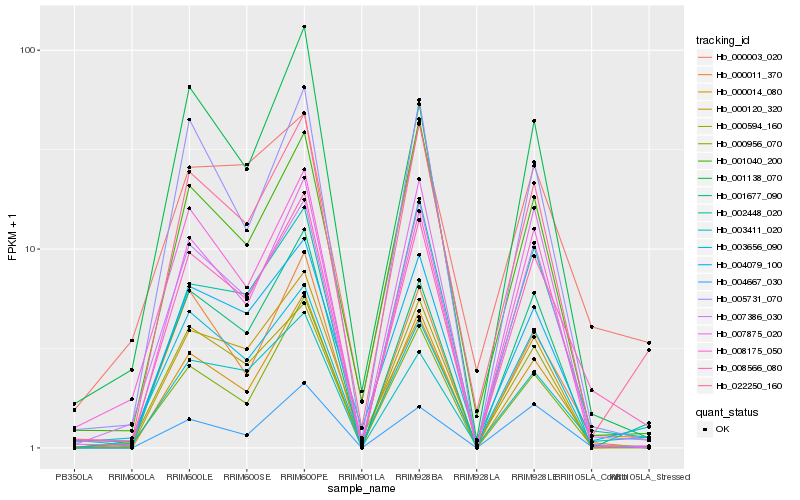
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008566_080 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647834 [Jatropha curcas] |
| 2 |
Hb_007386_030 |
0.0985912834 |
- |
- |
PREDICTED: formin-like protein 2 [Jatropha curcas] |
| 3 |
Hb_004079_100 |
0.1023484462 |
- |
- |
PREDICTED: uncharacterized protein LOC105637705 [Jatropha curcas] |
| 4 |
Hb_000011_370 |
0.1043474975 |
- |
- |
PREDICTED: uncharacterized protein LOC105631333 [Jatropha curcas] |
| 5 |
Hb_000014_080 |
0.1053282585 |
- |
- |
PREDICTED: UDP-glycosyltransferase 87A1-like [Jatropha curcas] |
| 6 |
Hb_022250_160 |
0.1062874686 |
- |
- |
respiratory burst oxidase A [Manihot esculenta] |
| 7 |
Hb_003411_020 |
0.1105990519 |
- |
- |
alpha-glucosidase, putative [Ricinus communis] |
| 8 |
Hb_000956_070 |
0.118039444 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g01540 [Jatropha curcas] |
| 9 |
Hb_001677_090 |
0.1184347326 |
- |
- |
PREDICTED: CASP-like protein 5B2 [Jatropha curcas] |
| 10 |
Hb_005731_070 |
0.121468139 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_004667_030 |
0.1225255822 |
- |
- |
hypothetical protein VITISV_034568 [Vitis vinifera] |
| 12 |
Hb_002448_020 |
0.1262135095 |
transcription factor |
TF Family: SBP |
Squamosa promoter-binding protein, putative [Ricinus communis] |
| 13 |
Hb_003656_090 |
0.1295602576 |
- |
- |
PREDICTED: protein Brevis radix-like 4 [Jatropha curcas] |
| 14 |
Hb_000120_320 |
0.1300966616 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 15 |
Hb_008175_050 |
0.1306613846 |
- |
- |
PREDICTED: formin-like protein 2 [Jatropha curcas] |
| 16 |
Hb_001040_200 |
0.1311422681 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA1H1-like [Jatropha curcas] |
| 17 |
Hb_000594_160 |
0.1343595788 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 8 [Jatropha curcas] |
| 18 |
Hb_001138_070 |
0.1354987581 |
- |
- |
PREDICTED: D-3-phosphoglycerate dehydrogenase 1, chloroplastic-like [Jatropha curcas] |
| 19 |
Hb_007875_020 |
0.1358471445 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme-like [Jatropha curcas] |
| 20 |
Hb_000003_020 |
0.1365081036 |
- |
- |
PREDICTED: uncharacterized protein LOC105631161 [Jatropha curcas] |