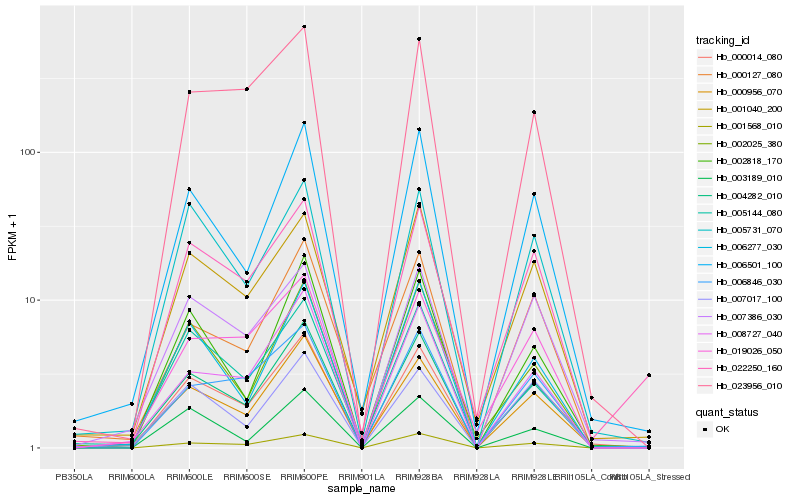
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004282_010 |
0.0 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At2g24230 [Jatropha curcas] |
| 2 |
Hb_000956_070 |
0.0513578646 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g01540 [Jatropha curcas] |
| 3 |
Hb_006501_100 |
0.0602461751 |
- |
- |
malic enzyme, putative [Ricinus communis] |
| 4 |
Hb_000014_080 |
0.0738134921 |
- |
- |
PREDICTED: UDP-glycosyltransferase 87A1-like [Jatropha curcas] |
| 5 |
Hb_008727_040 |
0.0874526992 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001568_010 |
0.0899475348 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_006277_030 |
0.0937200472 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_002818_170 |
0.0948998918 |
- |
- |
hypothetical protein POPTR_0014s10650g [Populus trichocarpa] |
| 9 |
Hb_000127_080 |
0.1027520548 |
- |
- |
hypothetical protein POPTR_0005s21760g [Populus trichocarpa] |
| 10 |
Hb_001040_200 |
0.1035536784 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA1H1-like [Jatropha curcas] |
| 11 |
Hb_005731_070 |
0.1069617562 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_005144_080 |
0.10869503 |
- |
- |
Serine carboxypeptidase, putative [Ricinus communis] |
| 13 |
Hb_002025_380 |
0.1166564474 |
- |
- |
Uncharacterized protein TCM_011176 [Theobroma cacao] |
| 14 |
Hb_007386_030 |
0.1179431918 |
- |
- |
PREDICTED: formin-like protein 2 [Jatropha curcas] |
| 15 |
Hb_007017_100 |
0.1191562172 |
- |
- |
PREDICTED: uncharacterized protein LOC105649804 isoform X1 [Jatropha curcas] |
| 16 |
Hb_019026_050 |
0.1227249582 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_023956_010 |
0.1232005949 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 18 |
Hb_006846_030 |
0.1239242697 |
- |
- |
respiratory burst oxidase A [Manihot esculenta] |
| 19 |
Hb_003189_010 |
0.1252784965 |
- |
- |
hypothetical protein EUGRSUZ_G02605 [Eucalyptus grandis] |
| 20 |
Hb_022250_160 |
0.1259543563 |
- |
- |
respiratory burst oxidase A [Manihot esculenta] |