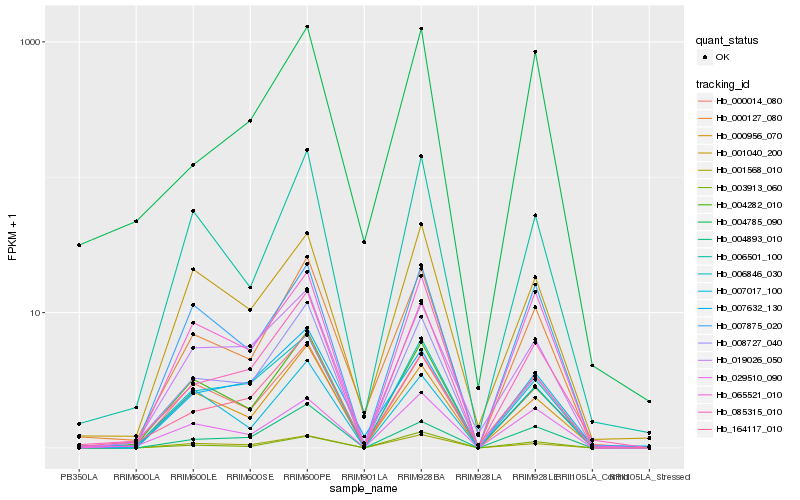
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000127_080 |
0.0 |
- |
- |
hypothetical protein POPTR_0005s21760g [Populus trichocarpa] |
| 2 |
Hb_004282_010 |
0.1027520548 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At2g24230 [Jatropha curcas] |
| 3 |
Hb_006501_100 |
0.105076379 |
- |
- |
malic enzyme, putative [Ricinus communis] |
| 4 |
Hb_000956_070 |
0.1084489573 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g01540 [Jatropha curcas] |
| 5 |
Hb_164117_010 |
0.1163739287 |
- |
- |
hypothetical protein PHAVU_004G0480000g [Phaseolus vulgaris] |
| 6 |
Hb_085315_010 |
0.1177971693 |
- |
- |
PREDICTED: uncharacterized protein LOC105632092 [Jatropha curcas] |
| 7 |
Hb_007632_130 |
0.123161057 |
- |
- |
PREDICTED: nudix hydrolase 10-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_008727_040 |
0.1264970173 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001040_200 |
0.128748299 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA1H1-like [Jatropha curcas] |
| 10 |
Hb_065521_010 |
0.1291905414 |
- |
- |
PREDICTED: probable metal-nicotianamine transporter YSL5 [Jatropha curcas] |
| 11 |
Hb_029510_090 |
0.1298465749 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At5g63020 [Jatropha curcas] |
| 12 |
Hb_001568_010 |
0.1301838783 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_007875_020 |
0.1313499064 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme-like [Jatropha curcas] |
| 14 |
Hb_000014_080 |
0.1329080856 |
- |
- |
PREDICTED: UDP-glycosyltransferase 87A1-like [Jatropha curcas] |
| 15 |
Hb_007017_100 |
0.138381339 |
- |
- |
PREDICTED: uncharacterized protein LOC105649804 isoform X1 [Jatropha curcas] |
| 16 |
Hb_004893_010 |
0.1391963569 |
- |
- |
PREDICTED: O-acyltransferase WSD1-like [Jatropha curcas] |
| 17 |
Hb_019026_050 |
0.1392699567 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_006846_030 |
0.1432283196 |
- |
- |
respiratory burst oxidase A [Manihot esculenta] |
| 19 |
Hb_004785_090 |
0.1454911971 |
- |
- |
hypothetical protein MIMGU_mgv1a018797mg [Erythranthe guttata] |
| 20 |
Hb_003913_060 |
0.149563597 |
- |
- |
ATP binding protein, putative [Ricinus communis] |