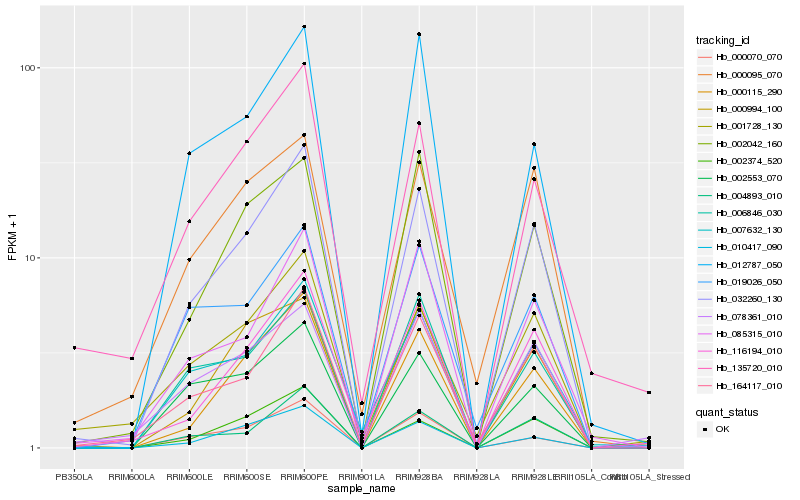
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_032260_130 |
0.0 |
- |
- |
hypothetical protein POPTR_0003s13800g [Populus trichocarpa] |
| 2 |
Hb_164117_010 |
0.0903410357 |
- |
- |
hypothetical protein PHAVU_004G0480000g [Phaseolus vulgaris] |
| 3 |
Hb_004893_010 |
0.09319209 |
- |
- |
PREDICTED: O-acyltransferase WSD1-like [Jatropha curcas] |
| 4 |
Hb_000115_290 |
0.0957432378 |
- |
- |
PREDICTED: uncharacterized protein LOC105124123 [Populus euphratica] |
| 5 |
Hb_116194_010 |
0.1011181404 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 6 |
Hb_007632_130 |
0.1040556267 |
- |
- |
PREDICTED: nudix hydrolase 10-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_002374_520 |
0.1069681723 |
- |
- |
early nodulin 8 precursor family protein [Populus trichocarpa] |
| 8 |
Hb_010417_090 |
0.117501528 |
- |
- |
PREDICTED: protein LURP-one-related 17 [Jatropha curcas] |
| 9 |
Hb_085315_010 |
0.1179300154 |
- |
- |
PREDICTED: uncharacterized protein LOC105632092 [Jatropha curcas] |
| 10 |
Hb_006846_030 |
0.1222375457 |
- |
- |
respiratory burst oxidase A [Manihot esculenta] |
| 11 |
Hb_002553_070 |
0.1243540215 |
- |
- |
PREDICTED: RING-H2 finger protein ATL16 [Jatropha curcas] |
| 12 |
Hb_000070_070 |
0.1261241324 |
- |
- |
PREDICTED: protein LURP-one-related 17 [Jatropha curcas] |
| 13 |
Hb_002042_160 |
0.1266305152 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 14 |
Hb_019026_050 |
0.1268683 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001728_130 |
0.1271572429 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 16 |
Hb_000994_100 |
0.1276195914 |
- |
- |
RALFL33, putative [Ricinus communis] |
| 17 |
Hb_135720_010 |
0.1292560543 |
- |
- |
hypothetical protein JCGZ_17857 [Jatropha curcas] |
| 18 |
Hb_000095_070 |
0.1314180685 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_012787_050 |
0.1316239373 |
- |
- |
PREDICTED: glutathione S-transferase U18-like [Jatropha curcas] |
| 20 |
Hb_078361_010 |
0.1353430652 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 1 [Jatropha curcas] |