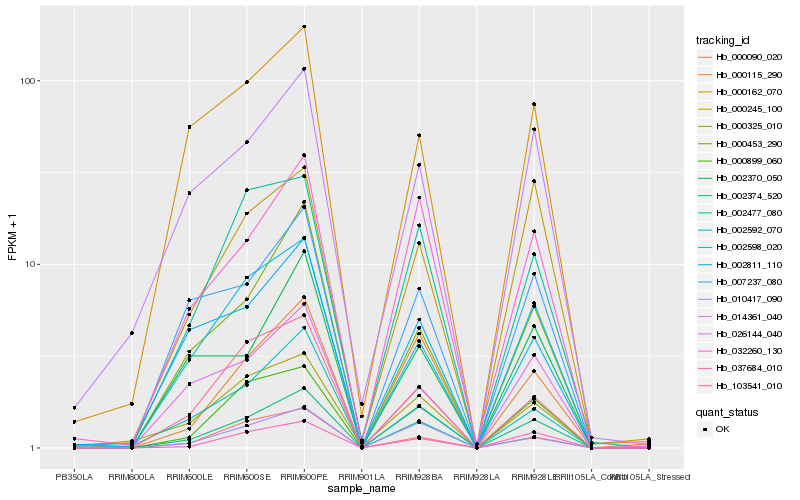
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002374_520 |
0.0 |
- |
- |
early nodulin 8 precursor family protein [Populus trichocarpa] |
| 2 |
Hb_000899_060 |
0.0954632119 |
- |
- |
PREDICTED: uncharacterized protein LOC105793631 [Gossypium raimondii] |
| 3 |
Hb_037684_010 |
0.1015997276 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105645015 [Jatropha curcas] |
| 4 |
Hb_002598_020 |
0.1066245425 |
- |
- |
PREDICTED: sugar transporter ERD6-like 5 [Populus euphratica] |
| 5 |
Hb_032260_130 |
0.1069681723 |
- |
- |
hypothetical protein POPTR_0003s13800g [Populus trichocarpa] |
| 6 |
Hb_007237_080 |
0.1229377649 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 7 |
Hb_000453_290 |
0.1246720351 |
- |
- |
PREDICTED: dr1-associated corepressor-like [Jatropha curcas] |
| 8 |
Hb_026144_040 |
0.1247252591 |
- |
- |
- |
| 9 |
Hb_000115_290 |
0.1262895609 |
- |
- |
PREDICTED: uncharacterized protein LOC105124123 [Populus euphratica] |
| 10 |
Hb_010417_090 |
0.1265270131 |
- |
- |
PREDICTED: protein LURP-one-related 17 [Jatropha curcas] |
| 11 |
Hb_014361_040 |
0.1293888075 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 12 |
Hb_002477_080 |
0.1319311454 |
- |
- |
hypothetical protein RCOM_0653450 [Ricinus communis] |
| 13 |
Hb_000090_020 |
0.132629377 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 14 |
Hb_002592_070 |
0.1361490518 |
- |
- |
hypothetical protein VITISV_020993 [Vitis vinifera] |
| 15 |
Hb_103541_010 |
0.136783272 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g63930 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000245_100 |
0.1377178451 |
- |
- |
PREDICTED: uncharacterized protein LOC105631555 [Jatropha curcas] |
| 17 |
Hb_000162_070 |
0.1394975761 |
- |
- |
phospholipase C, putative [Ricinus communis] |
| 18 |
Hb_000325_010 |
0.1406426123 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RGA2, putative [Ricinus communis] |
| 19 |
Hb_002370_050 |
0.1447987663 |
- |
- |
hypothetical protein JCGZ_18849 [Jatropha curcas] |
| 20 |
Hb_002811_110 |
0.1479078069 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK9 [Jatropha curcas] |