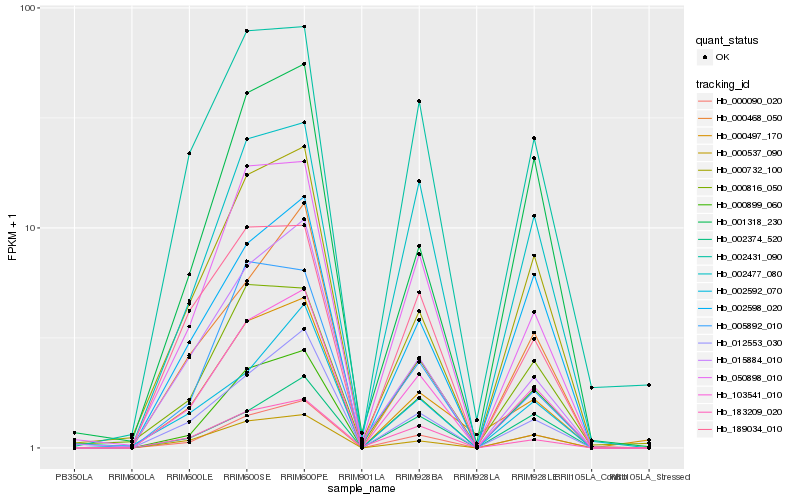
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_103541_010 |
0.0 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g63930 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000090_020 |
0.0356921944 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 3 |
Hb_012553_030 |
0.079797043 |
- |
- |
TRANSPARENT TESTA 12 protein, putative [Ricinus communis] |
| 4 |
Hb_050898_010 |
0.0942361765 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |
| 5 |
Hb_183209_020 |
0.1012716699 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g63930 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000537_090 |
0.1035842567 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_002598_020 |
0.1064633291 |
- |
- |
PREDICTED: sugar transporter ERD6-like 5 [Populus euphratica] |
| 8 |
Hb_002477_080 |
0.1101327721 |
- |
- |
hypothetical protein RCOM_0653450 [Ricinus communis] |
| 9 |
Hb_015884_010 |
0.1117780704 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g63930 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000732_100 |
0.1129541973 |
- |
- |
PREDICTED: uncharacterized protein LOC105649440 [Jatropha curcas] |
| 11 |
Hb_000468_050 |
0.1182070063 |
- |
- |
- |
| 12 |
Hb_002592_070 |
0.121128677 |
- |
- |
hypothetical protein VITISV_020993 [Vitis vinifera] |
| 13 |
Hb_002431_090 |
0.1232988284 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000899_060 |
0.1237032731 |
- |
- |
PREDICTED: uncharacterized protein LOC105793631 [Gossypium raimondii] |
| 15 |
Hb_000816_050 |
0.1238957897 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 16 |
Hb_000497_170 |
0.1279044984 |
- |
- |
PREDICTED: uncharacterized protein LOC105647275 [Jatropha curcas] |
| 17 |
Hb_189034_010 |
0.1282428101 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 18 |
Hb_001318_230 |
0.1309863357 |
- |
- |
Boron transporter, putative [Ricinus communis] |
| 19 |
Hb_005892_010 |
0.1330501363 |
- |
- |
PREDICTED: uncharacterized protein LOC105632168 [Jatropha curcas] |
| 20 |
Hb_002374_520 |
0.136783272 |
- |
- |
early nodulin 8 precursor family protein [Populus trichocarpa] |