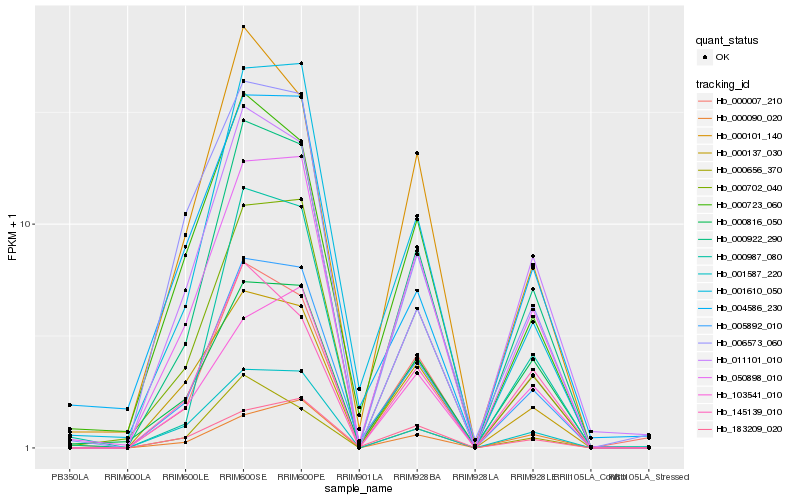
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005892_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632168 [Jatropha curcas] |
| 2 |
Hb_000922_290 |
0.0630862305 |
- |
- |
PREDICTED: probable boron transporter 2 [Jatropha curcas] |
| 3 |
Hb_050898_010 |
0.0763627692 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |
| 4 |
Hb_001610_050 |
0.0997049247 |
- |
- |
PREDICTED: subtilisin-like protease-like [Solanum tuberosum] |
| 5 |
Hb_001587_220 |
0.1078096569 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 6 |
Hb_000137_030 |
0.1088636886 |
- |
- |
PREDICTED: cysteine-rich repeat secretory protein 55-like [Citrus sinensis] |
| 7 |
Hb_000702_040 |
0.1104725617 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 8 |
Hb_000007_210 |
0.1134276403 |
- |
- |
hypothetical protein POPTR_0010s14360g [Populus trichocarpa] |
| 9 |
Hb_000987_080 |
0.1146036867 |
- |
- |
light-inducible protein atls1, putative [Ricinus communis] |
| 10 |
Hb_000723_060 |
0.1222498338 |
- |
- |
PREDICTED: uncharacterized protein LOC105642243 [Jatropha curcas] |
| 11 |
Hb_000816_050 |
0.1301310504 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 12 |
Hb_183209_020 |
0.1301653144 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g63930 isoform X2 [Jatropha curcas] |
| 13 |
Hb_103541_010 |
0.1330501363 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g63930 isoform X2 [Jatropha curcas] |
| 14 |
Hb_006573_060 |
0.1351582385 |
- |
- |
hypothetical protein [Bruguiera gymnorhiza] |
| 15 |
Hb_011101_010 |
0.1383311434 |
- |
- |
hypothetical protein JCGZ_25132 [Jatropha curcas] |
| 16 |
Hb_000101_140 |
0.1387748571 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 17 |
Hb_004586_230 |
0.1390254571 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000656_370 |
0.1404406933 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 19 |
Hb_000090_020 |
0.142619742 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 20 |
Hb_145139_010 |
0.1456416429 |
- |
- |
PREDICTED: sugar transporter ERD6-like 5 isoform X1 [Vitis vinifera] |