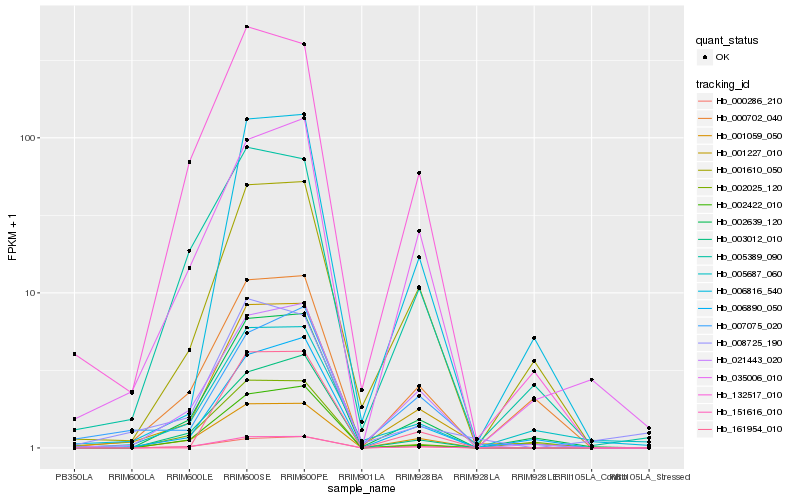
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001227_010 |
0.0 |
transcription factor |
TF Family: TCP |
hypothetical protein POPTR_0015s03140g [Populus trichocarpa] |
| 2 |
Hb_002639_120 |
0.0955275998 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 3 |
Hb_021443_020 |
0.1016221877 |
- |
- |
PREDICTED: uncharacterized protein LOC105648569 [Jatropha curcas] |
| 4 |
Hb_151616_010 |
0.1030649732 |
- |
- |
- |
| 5 |
Hb_006890_050 |
0.1092100972 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor CYCLOIDEA [Jatropha curcas] |
| 6 |
Hb_132517_010 |
0.112139029 |
- |
- |
PREDICTED: non-functional NADPH-dependent codeinone reductase 2-like isoform X3 [Populus euphratica] |
| 7 |
Hb_001610_050 |
0.1169791865 |
- |
- |
PREDICTED: subtilisin-like protease-like [Solanum tuberosum] |
| 8 |
Hb_000286_210 |
0.1178033523 |
- |
- |
hypothetical protein EUGRSUZ_C00018 [Eucalyptus grandis] |
| 9 |
Hb_001059_050 |
0.1210194869 |
- |
- |
PREDICTED: putative F-box/LRR-repeat protein At3g18150 [Jatropha curcas] |
| 10 |
Hb_006816_540 |
0.1244739051 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET16-like [Jatropha curcas] |
| 11 |
Hb_003012_010 |
0.1341836725 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_007075_020 |
0.1353076815 |
transcription factor |
TF Family: TRAF |
ankyrin repeat family protein [Populus trichocarpa] |
| 13 |
Hb_035006_010 |
0.1354333305 |
- |
- |
hypothetical protein POPTR_0017s11350g [Populus trichocarpa] |
| 14 |
Hb_000702_040 |
0.135727047 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 15 |
Hb_002422_010 |
0.1381426417 |
- |
- |
E3 ubiquitin-protein ligase UPL5 [Theobroma cacao] |
| 16 |
Hb_005389_090 |
0.1394755548 |
transcription factor |
TF Family: AUX/IAA |
Auxin-responsive protein IAA27, putative [Ricinus communis] |
| 17 |
Hb_005687_060 |
0.1450287691 |
- |
- |
PREDICTED: cinnamoyl-CoA reductase 1-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_002025_120 |
0.1499182325 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105634212 [Jatropha curcas] |
| 19 |
Hb_161954_010 |
0.1515432982 |
- |
- |
hypothetical protein JCGZ_05404 [Jatropha curcas] |
| 20 |
Hb_008725_190 |
0.1534924684 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein 5-like [Jatropha curcas] |