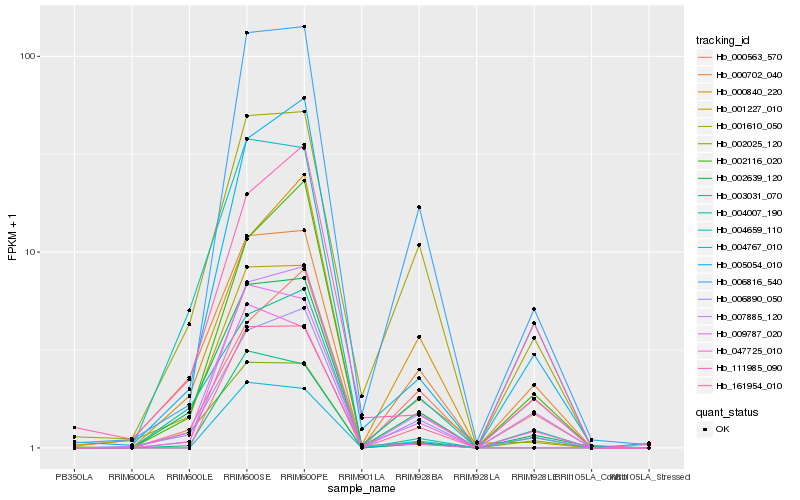
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007885_120 |
0.0 |
- |
- |
PREDICTED: BON1-associated protein 2-like [Jatropha curcas] |
| 2 |
Hb_006816_540 |
0.0898295551 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET16-like [Jatropha curcas] |
| 3 |
Hb_006890_050 |
0.0952255976 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor CYCLOIDEA [Jatropha curcas] |
| 4 |
Hb_005054_010 |
0.1050438229 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 5 |
Hb_003031_070 |
0.1118502397 |
- |
- |
PREDICTED: cysteine-rich repeat secretory protein 55-like [Jatropha curcas] |
| 6 |
Hb_009787_020 |
0.1204848986 |
- |
- |
PREDICTED: cytochrome P450 705A5-like [Jatropha curcas] |
| 7 |
Hb_002639_120 |
0.1298846616 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 8 |
Hb_000702_040 |
0.1427792408 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 9 |
Hb_000563_570 |
0.1477116119 |
- |
- |
hypothetical protein JCGZ_18895 [Jatropha curcas] |
| 10 |
Hb_001610_050 |
0.1491406396 |
- |
- |
PREDICTED: subtilisin-like protease-like [Solanum tuberosum] |
| 11 |
Hb_161954_010 |
0.1510603806 |
- |
- |
hypothetical protein JCGZ_05404 [Jatropha curcas] |
| 12 |
Hb_002025_120 |
0.1513261012 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105634212 [Jatropha curcas] |
| 13 |
Hb_000840_220 |
0.1539294712 |
- |
- |
PREDICTED: RING-H2 finger protein ATL78-like [Jatropha curcas] |
| 14 |
Hb_047725_010 |
0.1553663216 |
- |
- |
ceramidase, putative [Ricinus communis] |
| 15 |
Hb_111985_090 |
0.1574992772 |
- |
- |
hypothetical protein RCOM_1595950 [Ricinus communis] |
| 16 |
Hb_004007_190 |
0.1582614426 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein AZF2-like [Jatropha curcas] |
| 17 |
Hb_002116_020 |
0.1589469816 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 35 [Jatropha curcas] |
| 18 |
Hb_001227_010 |
0.1595900117 |
transcription factor |
TF Family: TCP |
hypothetical protein POPTR_0015s03140g [Populus trichocarpa] |
| 19 |
Hb_004659_110 |
0.1604908142 |
- |
- |
PREDICTED: BURP domain-containing protein 3-like [Jatropha curcas] |
| 20 |
Hb_004767_010 |
0.1622850916 |
- |
- |
hypothetical protein CICLE_v10010736mg [Citrus clementina] |