| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
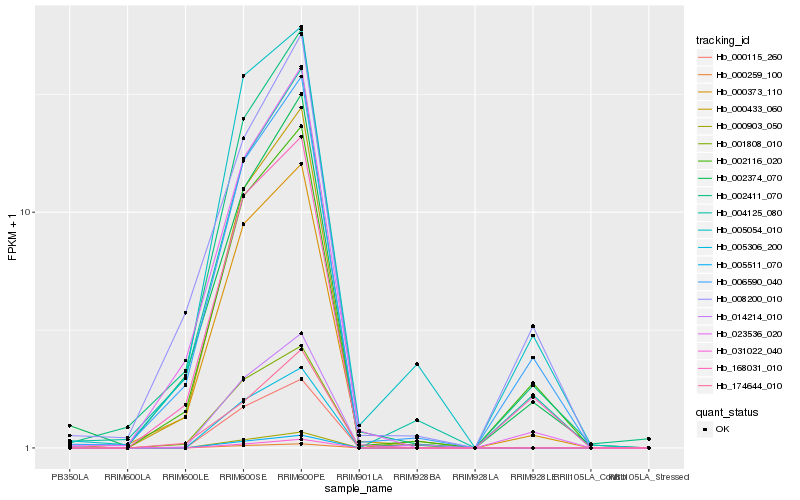
Hb_000373_110 |
0.0 |
- |
- |
Nodulin, putative [Ricinus communis] |
| 2 |
Hb_002411_070 |
0.0650939409 |
- |
- |
PREDICTED: WAT1-related protein At5g07050 [Jatropha curcas] |
| 3 |
Hb_168031_010 |
0.0703822593 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.6 [Jatropha curcas] |
| 4 |
Hb_023536_020 |
0.0720643536 |
- |
- |
Nicotianamine synthase, putative [Ricinus communis] |
| 5 |
Hb_004125_080 |
0.0833966575 |
- |
- |
PREDICTED: dCTP pyrophosphatase 1-like [Populus euphratica] |
| 6 |
Hb_002116_020 |
0.0836830841 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 35 [Jatropha curcas] |
| 7 |
Hb_000433_060 |
0.0839055809 |
- |
- |
hypothetical protein EUGRSUZ_B00708, partial [Eucalyptus grandis] |
| 8 |
Hb_006590_040 |
0.088606069 |
- |
- |
amino acid permease, putative [Ricinus communis] |
| 9 |
Hb_005054_010 |
0.0956938936 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 10 |
Hb_001808_010 |
0.097394085 |
- |
- |
PREDICTED: dirigent protein 23-like [Jatropha curcas] |
| 11 |
Hb_002374_070 |
0.0996184771 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 12 |
Hb_174644_010 |
0.10885134 |
- |
- |
- |
| 13 |
Hb_000115_260 |
0.1145622653 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH75 [Vitis vinifera] |
| 14 |
Hb_005511_070 |
0.1146280136 |
- |
- |
- |
| 15 |
Hb_005306_200 |
0.1149300681 |
- |
- |
- |
| 16 |
Hb_000903_050 |
0.115200522 |
- |
- |
hypothetical protein A259_26892 [Pseudomonas syringae pv. actinidiae ICMP 19070] |
| 17 |
Hb_000259_100 |
0.1158876629 |
- |
- |
hypothetical protein CICLE_v10033628mg, partial [Citrus clementina] |
| 18 |
Hb_014214_010 |
0.1164000441 |
- |
- |
PREDICTED: cationic amino acid transporter 6, chloroplastic-like [Jatropha curcas] |
| 19 |
Hb_031022_040 |
0.120661972 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Beta vulgaris subsp. vulgaris] |
| 20 |
Hb_008200_010 |
0.1260601921 |
- |
- |
PREDICTED: ABC transporter G family member 14-like isoform X1 [Jatropha curcas] |