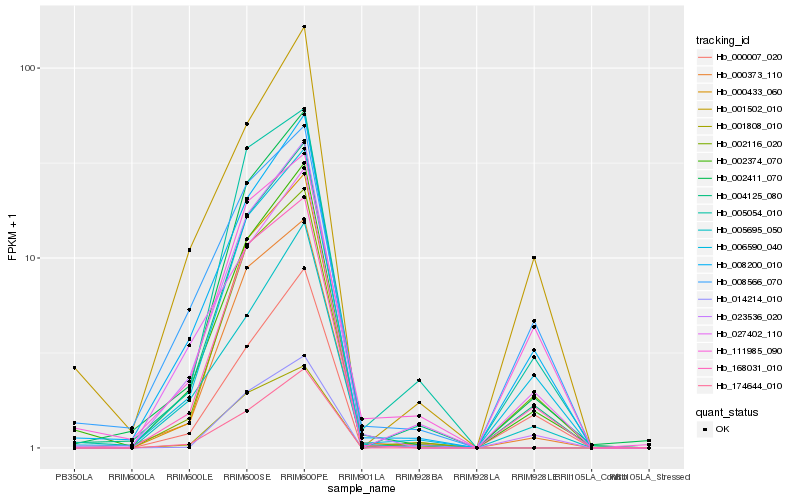
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002116_020 |
0.0 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 35 [Jatropha curcas] |
| 2 |
Hb_006590_040 |
0.0330591455 |
- |
- |
amino acid permease, putative [Ricinus communis] |
| 3 |
Hb_168031_010 |
0.0460250651 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.6 [Jatropha curcas] |
| 4 |
Hb_004125_080 |
0.0742249943 |
- |
- |
PREDICTED: dCTP pyrophosphatase 1-like [Populus euphratica] |
| 5 |
Hb_005054_010 |
0.0802576247 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 6 |
Hb_002411_070 |
0.0806719236 |
- |
- |
PREDICTED: WAT1-related protein At5g07050 [Jatropha curcas] |
| 7 |
Hb_000373_110 |
0.0836830841 |
- |
- |
Nodulin, putative [Ricinus communis] |
| 8 |
Hb_002374_070 |
0.0884354578 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 9 |
Hb_008200_010 |
0.0930088202 |
- |
- |
PREDICTED: ABC transporter G family member 14-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_000007_020 |
0.099770858 |
transcription factor |
TF Family: bZIP |
Ocs element-binding factor, putative [Ricinus communis] |
| 11 |
Hb_023536_020 |
0.1100973941 |
- |
- |
Nicotianamine synthase, putative [Ricinus communis] |
| 12 |
Hb_111985_090 |
0.113144952 |
- |
- |
hypothetical protein RCOM_1595950 [Ricinus communis] |
| 13 |
Hb_000433_060 |
0.1192986606 |
- |
- |
hypothetical protein EUGRSUZ_B00708, partial [Eucalyptus grandis] |
| 14 |
Hb_027402_110 |
0.1284178438 |
- |
- |
PREDICTED: uncharacterized protein LOC105640545 [Jatropha curcas] |
| 15 |
Hb_008566_070 |
0.1294603984 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001808_010 |
0.1329828943 |
- |
- |
PREDICTED: dirigent protein 23-like [Jatropha curcas] |
| 17 |
Hb_001502_010 |
0.1358014857 |
- |
- |
hypothetical protein JCGZ_04288 [Jatropha curcas] |
| 18 |
Hb_005695_050 |
0.1359164322 |
- |
- |
PREDICTED: sulfate transporter 1.3-like [Jatropha curcas] |
| 19 |
Hb_174644_010 |
0.1371657579 |
- |
- |
- |
| 20 |
Hb_014214_010 |
0.1386153085 |
- |
- |
PREDICTED: cationic amino acid transporter 6, chloroplastic-like [Jatropha curcas] |