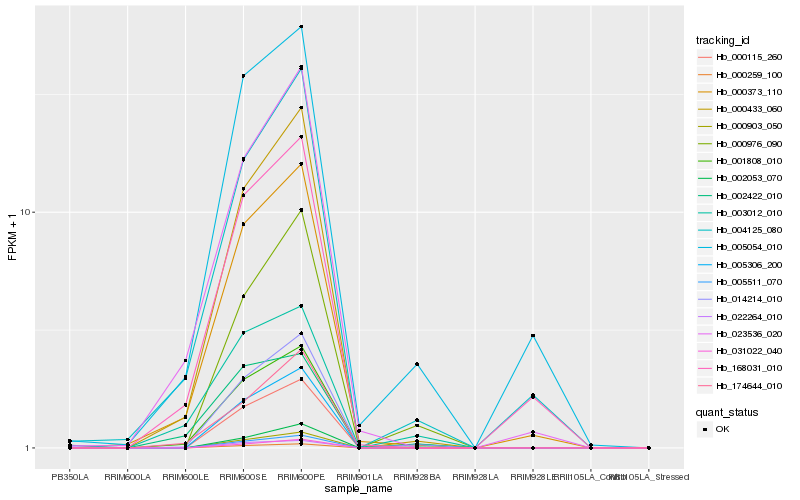
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001808_010 |
0.0 |
- |
- |
PREDICTED: dirigent protein 23-like [Jatropha curcas] |
| 2 |
Hb_014214_010 |
0.0755636201 |
- |
- |
PREDICTED: cationic amino acid transporter 6, chloroplastic-like [Jatropha curcas] |
| 3 |
Hb_000433_060 |
0.0803792071 |
- |
- |
hypothetical protein EUGRSUZ_B00708, partial [Eucalyptus grandis] |
| 4 |
Hb_000373_110 |
0.097394085 |
- |
- |
Nodulin, putative [Ricinus communis] |
| 5 |
Hb_002422_010 |
0.1092764109 |
- |
- |
E3 ubiquitin-protein ligase UPL5 [Theobroma cacao] |
| 6 |
Hb_003012_010 |
0.1108128778 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_005054_010 |
0.1112627508 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 8 |
Hb_002053_070 |
0.1129057765 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 14 [Populus euphratica] |
| 9 |
Hb_004125_080 |
0.1138231304 |
- |
- |
PREDICTED: dCTP pyrophosphatase 1-like [Populus euphratica] |
| 10 |
Hb_000976_090 |
0.1159810364 |
- |
- |
- |
| 11 |
Hb_000259_100 |
0.1171013424 |
- |
- |
hypothetical protein CICLE_v10033628mg, partial [Citrus clementina] |
| 12 |
Hb_000115_260 |
0.1178239274 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH75 [Vitis vinifera] |
| 13 |
Hb_005511_070 |
0.1180329776 |
- |
- |
- |
| 14 |
Hb_005306_200 |
0.1187334072 |
- |
- |
- |
| 15 |
Hb_000903_050 |
0.1192439106 |
- |
- |
hypothetical protein A259_26892 [Pseudomonas syringae pv. actinidiae ICMP 19070] |
| 16 |
Hb_174644_010 |
0.1223906721 |
- |
- |
- |
| 17 |
Hb_023536_020 |
0.1247214422 |
- |
- |
Nicotianamine synthase, putative [Ricinus communis] |
| 18 |
Hb_031022_040 |
0.1267098654 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Beta vulgaris subsp. vulgaris] |
| 19 |
Hb_022264_010 |
0.1272321582 |
- |
- |
PREDICTED: uncharacterized protein LOC105795741 [Gossypium raimondii] |
| 20 |
Hb_168031_010 |
0.1321478466 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.6 [Jatropha curcas] |