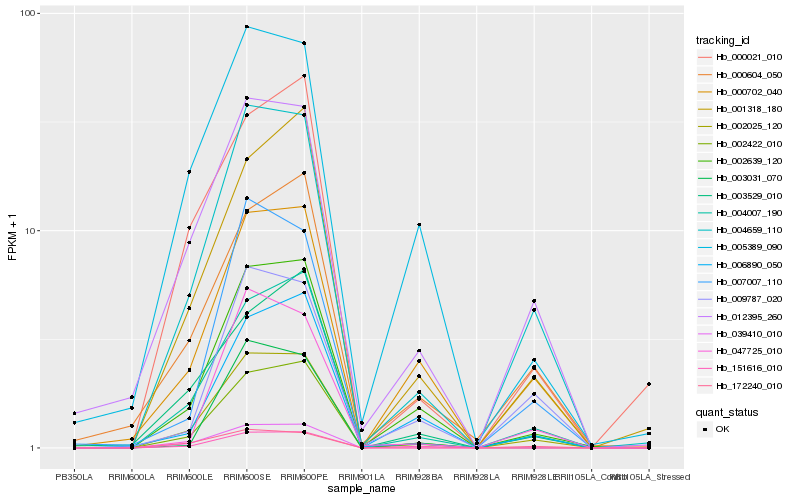
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002025_120 |
0.0 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105634212 [Jatropha curcas] |
| 2 |
Hb_004659_110 |
0.0561007056 |
- |
- |
PREDICTED: BURP domain-containing protein 3-like [Jatropha curcas] |
| 3 |
Hb_004007_190 |
0.0785959678 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein AZF2-like [Jatropha curcas] |
| 4 |
Hb_002639_120 |
0.0904931137 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 5 |
Hb_012395_260 |
0.1177932509 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA13 isoform X1 [Jatropha curcas] |
| 6 |
Hb_002422_010 |
0.120436411 |
- |
- |
E3 ubiquitin-protein ligase UPL5 [Theobroma cacao] |
| 7 |
Hb_000702_040 |
0.121974021 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 8 |
Hb_001318_180 |
0.1258960515 |
- |
- |
PREDICTED: protein NIM1-INTERACTING 1 [Jatropha curcas] |
| 9 |
Hb_003031_070 |
0.1335638629 |
- |
- |
PREDICTED: cysteine-rich repeat secretory protein 55-like [Jatropha curcas] |
| 10 |
Hb_151616_010 |
0.1342662691 |
- |
- |
- |
| 11 |
Hb_172240_010 |
0.1357797573 |
- |
- |
PREDICTED: uncharacterized protein LOC105795660 [Gossypium raimondii] |
| 12 |
Hb_000604_050 |
0.1359833073 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 8-like [Jatropha curcas] |
| 13 |
Hb_007007_110 |
0.1392310067 |
- |
- |
hypothetical protein JCGZ_14479 [Jatropha curcas] |
| 14 |
Hb_000021_010 |
0.139773555 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_006890_050 |
0.1398766476 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor CYCLOIDEA [Jatropha curcas] |
| 16 |
Hb_009787_020 |
0.1409879219 |
- |
- |
PREDICTED: cytochrome P450 705A5-like [Jatropha curcas] |
| 17 |
Hb_003529_010 |
0.1422702683 |
- |
- |
PREDICTED: disease resistance protein RPS6-like [Jatropha curcas] |
| 18 |
Hb_039410_010 |
0.1427701808 |
- |
- |
hypothetical protein JCGZ_23771 [Jatropha curcas] |
| 19 |
Hb_005389_090 |
0.1441244765 |
transcription factor |
TF Family: AUX/IAA |
Auxin-responsive protein IAA27, putative [Ricinus communis] |
| 20 |
Hb_047725_010 |
0.1441511046 |
- |
- |
ceramidase, putative [Ricinus communis] |