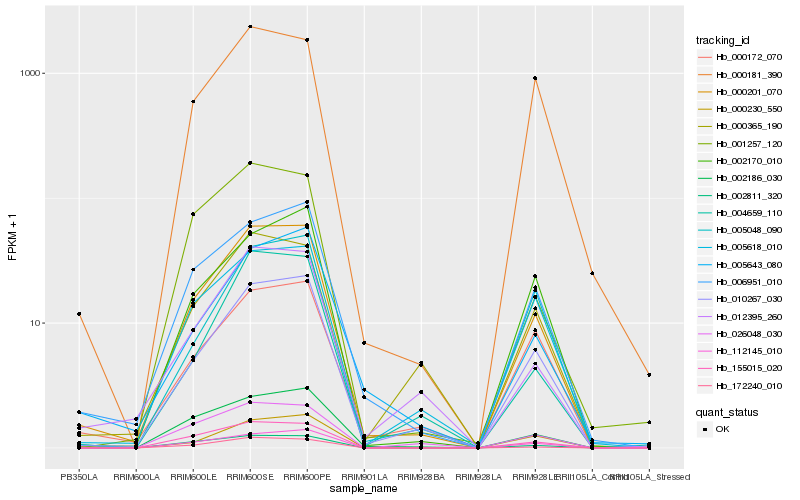
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000201_070 |
0.0 |
- |
- |
hypothetical protein POPTR_0067s00200g [Populus trichocarpa] |
| 2 |
Hb_010267_030 |
0.0726766117 |
- |
- |
DUF26 domain-containing protein 1 precursor, putative [Ricinus communis] |
| 3 |
Hb_006951_010 |
0.0969686514 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 10-like [Populus euphratica] |
| 4 |
Hb_112145_010 |
0.1006498701 |
- |
- |
kinase, putative [Ricinus communis] |
| 5 |
Hb_155015_020 |
0.1014290563 |
- |
- |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 6 |
Hb_172240_010 |
0.1085166572 |
- |
- |
PREDICTED: uncharacterized protein LOC105795660 [Gossypium raimondii] |
| 7 |
Hb_002186_030 |
0.1099110322 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g53430 isoform X1 [Populus euphratica] |
| 8 |
Hb_012395_260 |
0.1145063639 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA13 isoform X1 [Jatropha curcas] |
| 9 |
Hb_002811_320 |
0.1163118151 |
- |
- |
cation-transporting atpase plant, putative [Ricinus communis] |
| 10 |
Hb_005618_010 |
0.1204295416 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0014s10680g [Populus trichocarpa] |
| 11 |
Hb_000172_070 |
0.1231124634 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 12 |
Hb_005643_080 |
0.1244276158 |
- |
- |
hypothetical protein POPTR_0001s12740g [Populus trichocarpa] |
| 13 |
Hb_004659_110 |
0.124853949 |
- |
- |
PREDICTED: BURP domain-containing protein 3-like [Jatropha curcas] |
| 14 |
Hb_000230_550 |
0.1257709362 |
- |
- |
PREDICTED: adenylate isopentenyltransferase 5, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000365_190 |
0.1286912837 |
- |
- |
PREDICTED: protein LURP-one-related 8 [Jatropha curcas] |
| 16 |
Hb_002170_010 |
0.130692412 |
- |
- |
hypothetical protein JCGZ_25046 [Jatropha curcas] |
| 17 |
Hb_001257_120 |
0.1325312882 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 9 [Jatropha curcas] |
| 18 |
Hb_000181_390 |
0.1359164658 |
- |
- |
unknown [Lotus japonicus] |
| 19 |
Hb_005048_090 |
0.1376923092 |
- |
- |
PREDICTED: glycine-rich cell wall structural protein 2 [Jatropha curcas] |
| 20 |
Hb_026048_030 |
0.1379699761 |
- |
- |
conserved hypothetical protein [Ricinus communis] |