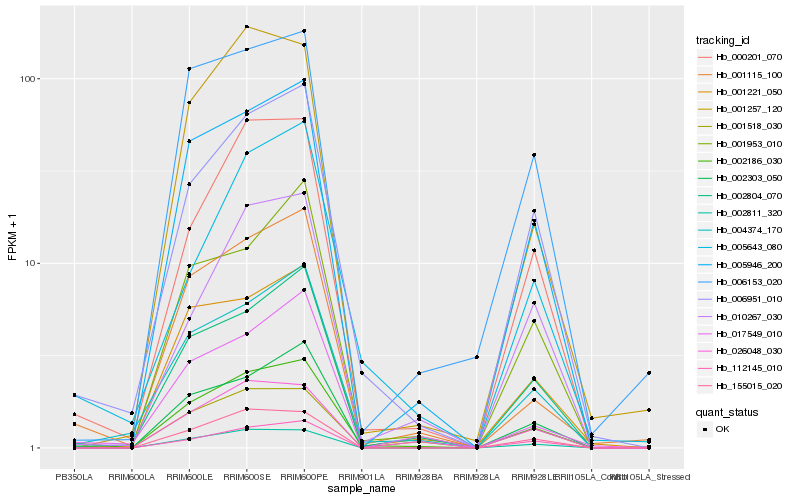
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002186_030 |
0.0 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g53430 isoform X1 [Populus euphratica] |
| 2 |
Hb_006951_010 |
0.0886315675 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 10-like [Populus euphratica] |
| 3 |
Hb_000201_070 |
0.1099110322 |
- |
- |
hypothetical protein POPTR_0067s00200g [Populus trichocarpa] |
| 4 |
Hb_004374_170 |
0.109919344 |
- |
- |
PREDICTED: gibberellin 20 oxidase 1-B [Jatropha curcas] |
| 5 |
Hb_005946_200 |
0.1122600807 |
- |
- |
hypothetical protein POPTR_0006s16210g [Populus trichocarpa] |
| 6 |
Hb_001115_100 |
0.1133482529 |
- |
- |
PREDICTED: uncharacterized protein LOC103714121 [Phoenix dactylifera] |
| 7 |
Hb_002804_070 |
0.1163785955 |
- |
- |
hypothetical protein JCGZ_05701 [Jatropha curcas] |
| 8 |
Hb_002811_320 |
0.1221596163 |
- |
- |
cation-transporting atpase plant, putative [Ricinus communis] |
| 9 |
Hb_155015_020 |
0.1269238148 |
- |
- |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 10 |
Hb_001221_050 |
0.1276928632 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At3g03770 [Jatropha curcas] |
| 11 |
Hb_112145_010 |
0.1303552099 |
- |
- |
kinase, putative [Ricinus communis] |
| 12 |
Hb_002303_050 |
0.1315776269 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor PTL-like [Populus euphratica] |
| 13 |
Hb_001518_030 |
0.1370642846 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 14 |
Hb_006153_020 |
0.1411532886 |
- |
- |
vacuolar invertase [Manihot esculenta] |
| 15 |
Hb_017549_010 |
0.1414556416 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_00222 [Jatropha curcas] |
| 16 |
Hb_001953_010 |
0.1421938997 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET10-like [Jatropha curcas] |
| 17 |
Hb_005643_080 |
0.1445684693 |
- |
- |
hypothetical protein POPTR_0001s12740g [Populus trichocarpa] |
| 18 |
Hb_026048_030 |
0.145125726 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001257_120 |
0.1466533212 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 9 [Jatropha curcas] |
| 20 |
Hb_010267_030 |
0.1480310198 |
- |
- |
DUF26 domain-containing protein 1 precursor, putative [Ricinus communis] |