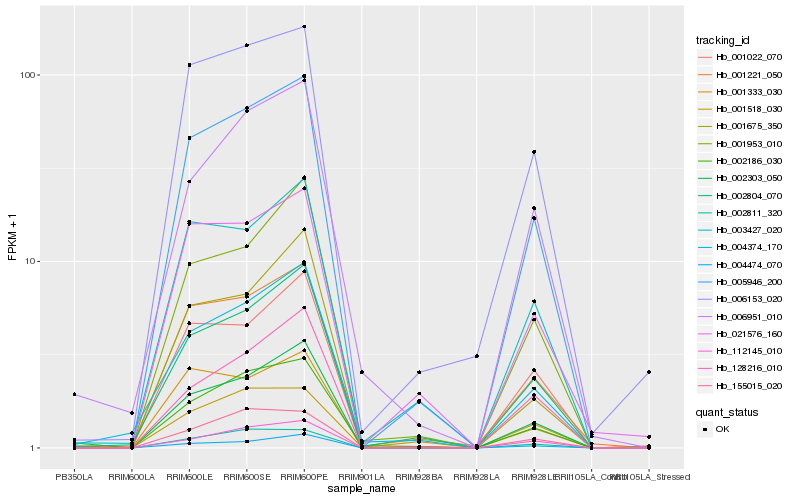
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005946_200 |
0.0 |
- |
- |
hypothetical protein POPTR_0006s16210g [Populus trichocarpa] |
| 2 |
Hb_001221_050 |
0.068685057 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At3g03770 [Jatropha curcas] |
| 3 |
Hb_002303_050 |
0.0713246712 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor PTL-like [Populus euphratica] |
| 4 |
Hb_002804_070 |
0.0756343396 |
- |
- |
hypothetical protein JCGZ_05701 [Jatropha curcas] |
| 5 |
Hb_006153_020 |
0.0839845841 |
- |
- |
vacuolar invertase [Manihot esculenta] |
| 6 |
Hb_001022_070 |
0.0901240428 |
- |
- |
cell wall invertase [Manihot esculenta] |
| 7 |
Hb_002811_320 |
0.0918668269 |
- |
- |
cation-transporting atpase plant, putative [Ricinus communis] |
| 8 |
Hb_004474_070 |
0.0942598148 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPP8, putative [Ricinus communis] |
| 9 |
Hb_003427_020 |
0.0954249205 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g53440 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001953_010 |
0.0986891551 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET10-like [Jatropha curcas] |
| 11 |
Hb_021576_160 |
0.1005085085 |
transcription factor |
TF Family: WRKY |
putative WRKY transcription factor family protein [Populus trichocarpa] |
| 12 |
Hb_112145_010 |
0.1028054984 |
- |
- |
kinase, putative [Ricinus communis] |
| 13 |
Hb_004374_170 |
0.1052906898 |
- |
- |
PREDICTED: gibberellin 20 oxidase 1-B [Jatropha curcas] |
| 14 |
Hb_001333_030 |
0.1063416738 |
- |
- |
Abscisic acid 8'-hydroxylase 1 [Theobroma cacao] |
| 15 |
Hb_155015_020 |
0.1078886548 |
- |
- |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 16 |
Hb_002186_030 |
0.1122600807 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g53430 isoform X1 [Populus euphratica] |
| 17 |
Hb_128216_010 |
0.1142427737 |
- |
- |
PREDICTED: UDP-glycosyltransferase 79B6-like [Jatropha curcas] |
| 18 |
Hb_001518_030 |
0.1200536019 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 19 |
Hb_001675_350 |
0.1219519033 |
- |
- |
Avr9/Cf-9 rapidly elicited protein [Jatropha curcas] |
| 20 |
Hb_006951_010 |
0.1228561785 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 10-like [Populus euphratica] |