| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
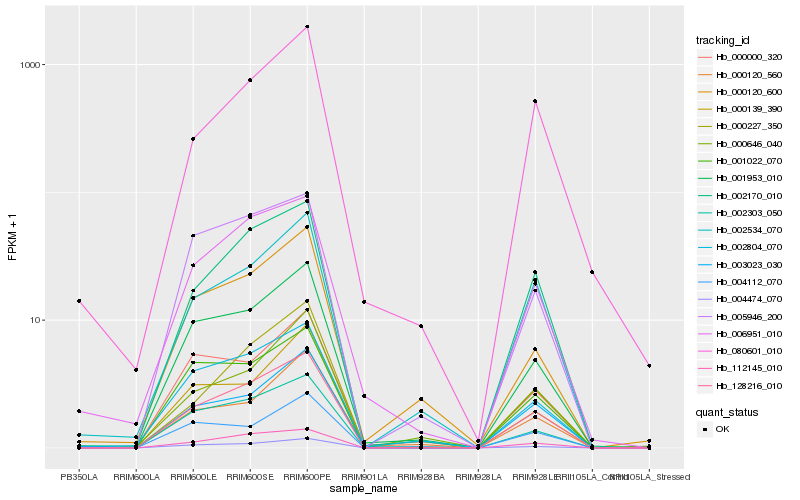
Hb_128216_010 |
0.0 |
- |
- |
PREDICTED: UDP-glycosyltransferase 79B6-like [Jatropha curcas] |
| 2 |
Hb_002170_010 |
0.0672098346 |
- |
- |
hypothetical protein JCGZ_25046 [Jatropha curcas] |
| 3 |
Hb_004474_070 |
0.0706013931 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPP8, putative [Ricinus communis] |
| 4 |
Hb_112145_010 |
0.0757826735 |
- |
- |
kinase, putative [Ricinus communis] |
| 5 |
Hb_002303_050 |
0.0809935999 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor PTL-like [Populus euphratica] |
| 6 |
Hb_001953_010 |
0.0830138514 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET10-like [Jatropha curcas] |
| 7 |
Hb_002804_070 |
0.0925460837 |
- |
- |
hypothetical protein JCGZ_05701 [Jatropha curcas] |
| 8 |
Hb_003023_030 |
0.096289269 |
- |
- |
PREDICTED: MATE efflux family protein 5-like isoform X1 [Citrus sinensis] |
| 9 |
Hb_002534_070 |
0.098688996 |
- |
- |
PREDICTED: uncharacterized protein LOC105640545 [Jatropha curcas] |
| 10 |
Hb_006951_010 |
0.1089880811 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 10-like [Populus euphratica] |
| 11 |
Hb_005946_200 |
0.1142427737 |
- |
- |
hypothetical protein POPTR_0006s16210g [Populus trichocarpa] |
| 12 |
Hb_000227_350 |
0.1153519066 |
- |
- |
hypothetical protein CICLE_v10006537mg [Citrus clementina] |
| 13 |
Hb_001022_070 |
0.1162117998 |
- |
- |
cell wall invertase [Manihot esculenta] |
| 14 |
Hb_000646_040 |
0.1198304026 |
- |
- |
PREDICTED: thiamine-repressible mitochondrial transport protein THI74-like [Jatropha curcas] |
| 15 |
Hb_000120_600 |
0.123587469 |
- |
- |
PREDICTED: uncharacterized protein LOC105628582 [Jatropha curcas] |
| 16 |
Hb_000139_390 |
0.1237993079 |
- |
- |
- |
| 17 |
Hb_000120_560 |
0.1247180737 |
- |
- |
protein with unknown function [Ricinus communis] |
| 18 |
Hb_004112_070 |
0.1250890358 |
- |
- |
- |
| 19 |
Hb_000000_320 |
0.1273489049 |
- |
- |
PREDICTED: calcium uniporter protein 4, mitochondrial [Jatropha curcas] |
| 20 |
Hb_080601_010 |
0.1286736357 |
- |
- |
metallothionein 3-like protein [Hevea brasiliensis] |