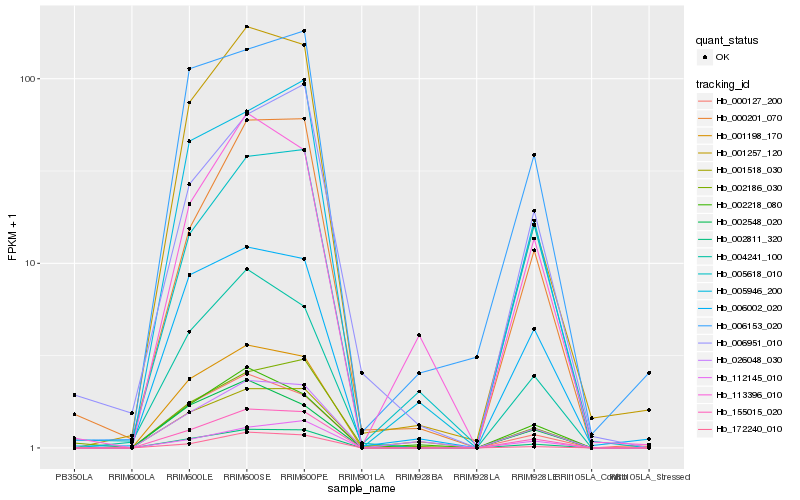
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_155015_020 |
0.0 |
- |
- |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 2 |
Hb_002811_320 |
0.0244063843 |
- |
- |
cation-transporting atpase plant, putative [Ricinus communis] |
| 3 |
Hb_001198_170 |
0.0832575914 |
- |
- |
- |
| 4 |
Hb_001257_120 |
0.0914616023 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 9 [Jatropha curcas] |
| 5 |
Hb_112145_010 |
0.0944182889 |
- |
- |
kinase, putative [Ricinus communis] |
| 6 |
Hb_172240_010 |
0.1004024771 |
- |
- |
PREDICTED: uncharacterized protein LOC105795660 [Gossypium raimondii] |
| 7 |
Hb_000201_070 |
0.1014290563 |
- |
- |
hypothetical protein POPTR_0067s00200g [Populus trichocarpa] |
| 8 |
Hb_004241_100 |
0.1062585219 |
- |
- |
PREDICTED: transcription factor bHLH123-like isoform X1 [Populus euphratica] |
| 9 |
Hb_005946_200 |
0.1078886548 |
- |
- |
hypothetical protein POPTR_0006s16210g [Populus trichocarpa] |
| 10 |
Hb_026048_030 |
0.1118393634 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002218_080 |
0.125500852 |
- |
- |
Ankyrin repeat family protein, putative [Theobroma cacao] |
| 12 |
Hb_002186_030 |
0.1269238148 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g53430 isoform X1 [Populus euphratica] |
| 13 |
Hb_001518_030 |
0.1276755752 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 14 |
Hb_006002_020 |
0.1278314048 |
- |
- |
PREDICTED: cytokinin dehydrogenase 7 [Jatropha curcas] |
| 15 |
Hb_006153_020 |
0.1285030624 |
- |
- |
vacuolar invertase [Manihot esculenta] |
| 16 |
Hb_002548_020 |
0.1291197809 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000127_200 |
0.1293249062 |
- |
- |
PREDICTED: uncharacterized protein LOC100255537 [Vitis vinifera] |
| 18 |
Hb_005618_010 |
0.1299733549 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0014s10680g [Populus trichocarpa] |
| 19 |
Hb_113396_010 |
0.1332632705 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 20 |
Hb_006951_010 |
0.1346401165 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 10-like [Populus euphratica] |