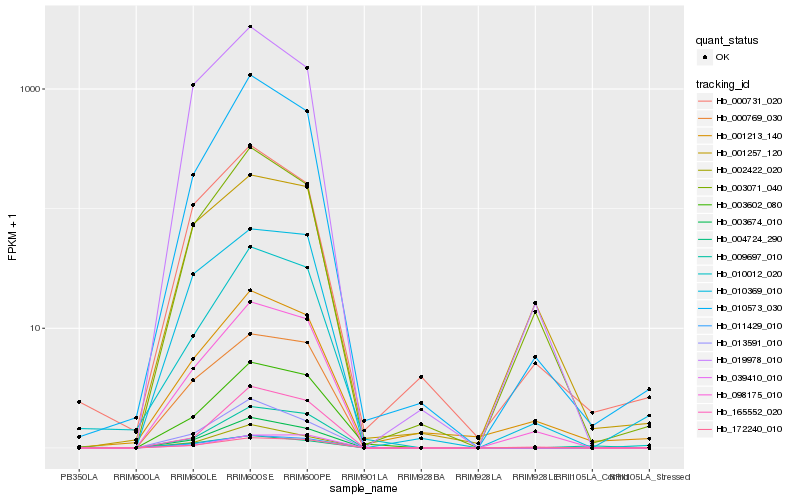
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_098175_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_23771 [Jatropha curcas] |
| 2 |
Hb_172240_010 |
0.0719535583 |
- |
- |
PREDICTED: uncharacterized protein LOC105795660 [Gossypium raimondii] |
| 3 |
Hb_003071_040 |
0.0848216773 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF017 [Jatropha curcas] |
| 4 |
Hb_011429_010 |
0.0861705123 |
- |
- |
hypothetical protein POPTR_0188s00230g [Populus trichocarpa] |
| 5 |
Hb_000769_030 |
0.0958012255 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 6 |
Hb_010573_030 |
0.1011964186 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_003602_080 |
0.1074713247 |
- |
- |
hypothetical protein POPTR_0002s10890g [Populus trichocarpa] |
| 8 |
Hb_001257_120 |
0.1079549341 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 9 [Jatropha curcas] |
| 9 |
Hb_010369_010 |
0.1104285805 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_003674_010 |
0.1168134939 |
- |
- |
purine transporter, putative [Ricinus communis] |
| 11 |
Hb_004724_290 |
0.1184117736 |
transcription factor |
TF Family: NAC |
- |
| 12 |
Hb_002422_020 |
0.1184960226 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL5 [Jatropha curcas] |
| 13 |
Hb_010012_020 |
0.119760646 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_019978_010 |
0.1221463136 |
- |
- |
PREDICTED: glycerophosphodiester phosphodiesterase protein kinase domain-containing GDPDL2-like isoform X2 [Jatropha curcas] |
| 15 |
Hb_013591_010 |
0.1231943296 |
- |
- |
Mitogen-activated protein kinase kinase kinase 1 [Glycine soja] |
| 16 |
Hb_039410_010 |
0.124183122 |
- |
- |
hypothetical protein JCGZ_23771 [Jatropha curcas] |
| 17 |
Hb_001213_140 |
0.12711282 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 18 |
Hb_165552_020 |
0.1271646704 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 19 |
Hb_000731_020 |
0.1290719706 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein ZAT10-like [Jatropha curcas] |
| 20 |
Hb_009697_010 |
0.1299771205 |
- |
- |
conserved hypothetical protein [Ricinus communis] |