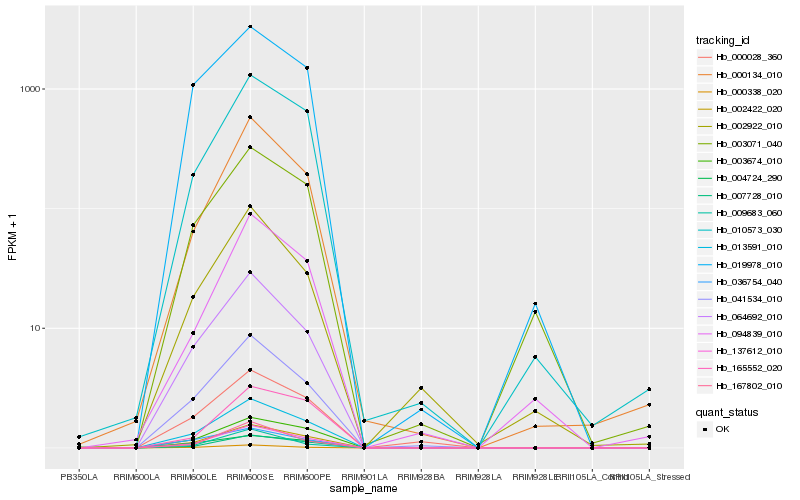
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_137612_010 |
0.0 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 2 |
Hb_002422_020 |
0.0368283796 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL5 [Jatropha curcas] |
| 3 |
Hb_013591_010 |
0.0371550745 |
- |
- |
Mitogen-activated protein kinase kinase kinase 1 [Glycine soja] |
| 4 |
Hb_007728_010 |
0.051980029 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Jatropha curcas] |
| 5 |
Hb_000134_010 |
0.0651998628 |
- |
- |
- |
| 6 |
Hb_041534_010 |
0.0673603732 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF017 [Jatropha curcas] |
| 7 |
Hb_064692_010 |
0.0673722632 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 8 |
Hb_010573_030 |
0.0733028361 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_167802_010 |
0.101008776 |
- |
- |
PREDICTED: receptor-like protein 12 [Populus euphratica] |
| 10 |
Hb_000338_020 |
0.1104991155 |
- |
- |
PREDICTED: tryptophan synthase beta chain 1 [Jatropha curcas] |
| 11 |
Hb_094839_010 |
0.1115456412 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_003674_010 |
0.1118242944 |
- |
- |
purine transporter, putative [Ricinus communis] |
| 13 |
Hb_019978_010 |
0.1137997103 |
- |
- |
PREDICTED: glycerophosphodiester phosphodiesterase protein kinase domain-containing GDPDL2-like isoform X2 [Jatropha curcas] |
| 14 |
Hb_000028_360 |
0.1201693438 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 15 |
Hb_002922_010 |
0.1234341173 |
- |
- |
heavy-metal-associated domain-containing family protein [Populus trichocarpa] |
| 16 |
Hb_003071_040 |
0.1269389735 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF017 [Jatropha curcas] |
| 17 |
Hb_165552_020 |
0.1289518754 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 18 |
Hb_004724_290 |
0.1294425985 |
transcription factor |
TF Family: NAC |
- |
| 19 |
Hb_009683_060 |
0.1294809986 |
- |
- |
- |
| 20 |
Hb_036754_040 |
0.1303304012 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |