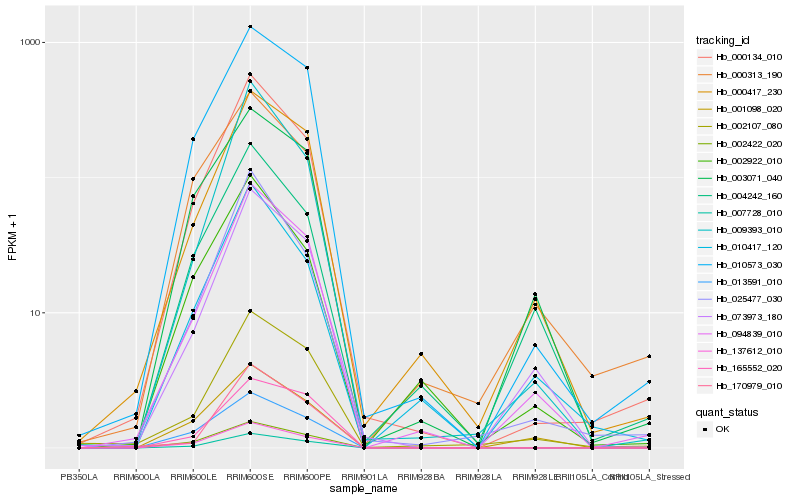
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_094839_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000417_230 |
0.0597564172 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_073973_180 |
0.0703739513 |
- |
- |
PREDICTED: WAT1-related protein At4g08290 isoform X1 [Populus euphratica] |
| 4 |
Hb_010573_030 |
0.085948763 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000134_010 |
0.086565484 |
- |
- |
- |
| 6 |
Hb_002107_080 |
0.0891210933 |
- |
- |
JHL20J20.3 [Jatropha curcas] |
| 7 |
Hb_007728_010 |
0.0949672871 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Jatropha curcas] |
| 8 |
Hb_010417_120 |
0.1069585274 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_009393_010 |
0.1079771116 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF017 [Jatropha curcas] |
| 10 |
Hb_137612_010 |
0.1115456412 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 11 |
Hb_004242_160 |
0.1140061487 |
transcription factor |
TF Family: GRAS |
PREDICTED: chitin-inducible gibberellin-responsive protein 1-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_003071_040 |
0.1164577292 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF017 [Jatropha curcas] |
| 13 |
Hb_025477_030 |
0.1165320155 |
transcription factor |
TF Family: ERF |
dehydration-responsive element-binding protein 2 [Manihot esculenta] |
| 14 |
Hb_001098_020 |
0.1228253755 |
- |
- |
PREDICTED: transcription factor bHLH123-like isoform X1 [Populus euphratica] |
| 15 |
Hb_002922_010 |
0.126678006 |
- |
- |
heavy-metal-associated domain-containing family protein [Populus trichocarpa] |
| 16 |
Hb_002422_020 |
0.1268309595 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL5 [Jatropha curcas] |
| 17 |
Hb_013591_010 |
0.1303295856 |
- |
- |
Mitogen-activated protein kinase kinase kinase 1 [Glycine soja] |
| 18 |
Hb_000313_190 |
0.1311013768 |
- |
- |
PREDICTED: uncharacterized protein LOC105636896 [Jatropha curcas] |
| 19 |
Hb_170979_010 |
0.132194201 |
- |
- |
PREDICTED: peroxidase 4-like [Jatropha curcas] |
| 20 |
Hb_165552_020 |
0.1343574316 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |