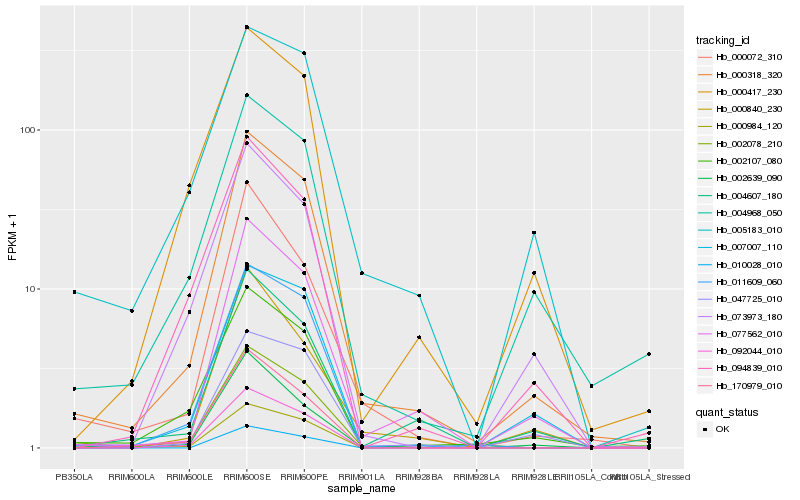
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000318_320 |
0.0 |
- |
- |
PREDICTED: transmembrane protein 45B [Jatropha curcas] |
| 2 |
Hb_077562_010 |
0.0998821442 |
- |
- |
PREDICTED: WAT1-related protein At5g07050 [Jatropha curcas] |
| 3 |
Hb_002107_080 |
0.1048736291 |
- |
- |
JHL20J20.3 [Jatropha curcas] |
| 4 |
Hb_000984_120 |
0.1056185355 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 5 |
Hb_007007_110 |
0.1115059753 |
- |
- |
hypothetical protein JCGZ_14479 [Jatropha curcas] |
| 6 |
Hb_002078_210 |
0.1130639192 |
- |
- |
PREDICTED: cytochrome P450 78A5-like [Jatropha curcas] |
| 7 |
Hb_011609_060 |
0.1144744848 |
- |
- |
PREDICTED: cytokinin hydroxylase [Jatropha curcas] |
| 8 |
Hb_092044_010 |
0.1175404917 |
- |
- |
PREDICTED: uncharacterized protein LOC105636336 [Jatropha curcas] |
| 9 |
Hb_000072_310 |
0.1186610592 |
transcription factor |
TF Family: bZIP |
PREDICTED: basic leucine zipper 43-like [Jatropha curcas] |
| 10 |
Hb_047725_010 |
0.1214552014 |
- |
- |
ceramidase, putative [Ricinus communis] |
| 11 |
Hb_002639_090 |
0.1220912559 |
- |
- |
hypothetical protein JCGZ_08134 [Jatropha curcas] |
| 12 |
Hb_004607_180 |
0.1246878599 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000417_230 |
0.1258517679 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_004968_050 |
0.1268972874 |
- |
- |
PREDICTED: putative lipid-transfer protein DIR1 [Jatropha curcas] |
| 15 |
Hb_170979_010 |
0.1294088159 |
- |
- |
PREDICTED: peroxidase 4-like [Jatropha curcas] |
| 16 |
Hb_073973_180 |
0.132799881 |
- |
- |
PREDICTED: WAT1-related protein At4g08290 isoform X1 [Populus euphratica] |
| 17 |
Hb_094839_010 |
0.1380911165 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_005183_010 |
0.1392999579 |
- |
- |
PREDICTED: probable calcium-binding protein CML17 [Malus domestica] |
| 19 |
Hb_000840_230 |
0.1393603921 |
- |
- |
RING-H2 finger protein ATL1L, putative [Ricinus communis] |
| 20 |
Hb_010028_010 |
0.1466553534 |
- |
- |
serine/threonine-protein kinase bri1, putative [Ricinus communis] |