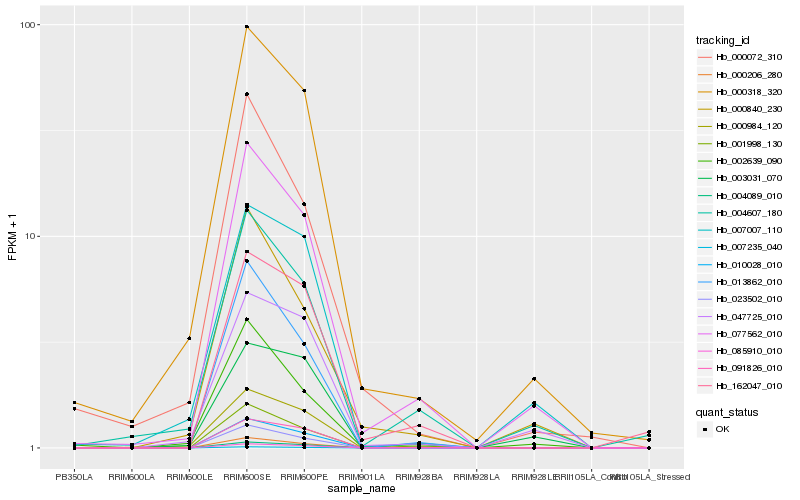
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_077562_010 |
0.0 |
- |
- |
PREDICTED: WAT1-related protein At5g07050 [Jatropha curcas] |
| 2 |
Hb_000318_320 |
0.0998821442 |
- |
- |
PREDICTED: transmembrane protein 45B [Jatropha curcas] |
| 3 |
Hb_004607_180 |
0.1012604579 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000840_230 |
0.1112743656 |
- |
- |
RING-H2 finger protein ATL1L, putative [Ricinus communis] |
| 5 |
Hb_002639_090 |
0.1121962711 |
- |
- |
hypothetical protein JCGZ_08134 [Jatropha curcas] |
| 6 |
Hb_001998_130 |
0.1142409083 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor 35 family protein [Populus trichocarpa] |
| 7 |
Hb_047725_010 |
0.124833628 |
- |
- |
ceramidase, putative [Ricinus communis] |
| 8 |
Hb_013862_010 |
0.1321637512 |
- |
- |
PREDICTED: uncharacterized protein LOC105650642 [Jatropha curcas] |
| 9 |
Hb_091826_010 |
0.1365235402 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_000984_120 |
0.136973225 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 11 |
Hb_007007_110 |
0.1404595063 |
- |
- |
hypothetical protein JCGZ_14479 [Jatropha curcas] |
| 12 |
Hb_003031_070 |
0.1426491658 |
- |
- |
PREDICTED: cysteine-rich repeat secretory protein 55-like [Jatropha curcas] |
| 13 |
Hb_000072_310 |
0.1456459493 |
transcription factor |
TF Family: bZIP |
PREDICTED: basic leucine zipper 43-like [Jatropha curcas] |
| 14 |
Hb_007235_040 |
0.1456963378 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 15 |
Hb_010028_010 |
0.1459681954 |
- |
- |
serine/threonine-protein kinase bri1, putative [Ricinus communis] |
| 16 |
Hb_000206_280 |
0.1463471028 |
- |
- |
Uncharacterized protein TCM_031897 [Theobroma cacao] |
| 17 |
Hb_023502_010 |
0.14668816 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 18 |
Hb_004089_010 |
0.148656391 |
- |
- |
hypothetical protein MIMGU_mgv1a018902mg [Erythranthe guttata] |
| 19 |
Hb_085910_010 |
0.1520750758 |
- |
- |
PREDICTED: uncharacterized protein LOC104106381 [Nicotiana tomentosiformis] |
| 20 |
Hb_162047_010 |
0.1535091493 |
- |
- |
PREDICTED: protein indeterminate-domain 16 [Jatropha curcas] |