| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
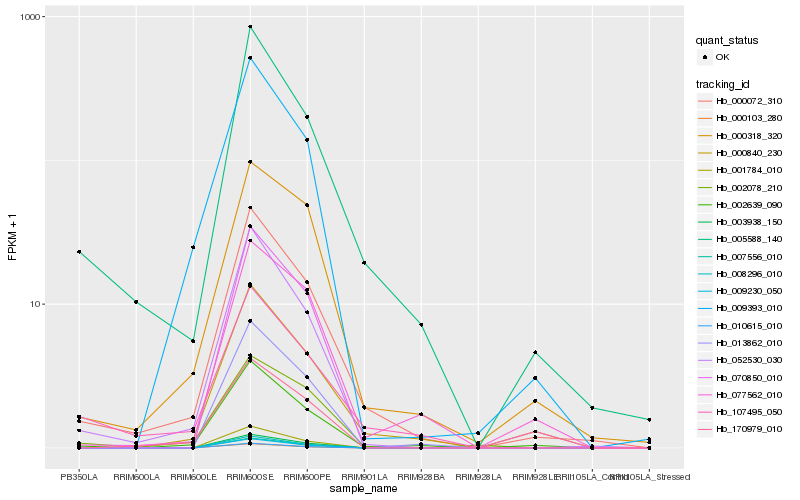
Hb_000072_310 |
0.0 |
transcription factor |
TF Family: bZIP |
PREDICTED: basic leucine zipper 43-like [Jatropha curcas] |
| 2 |
Hb_005588_140 |
0.071890937 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002639_090 |
0.0983375094 |
- |
- |
hypothetical protein JCGZ_08134 [Jatropha curcas] |
| 4 |
Hb_052530_030 |
0.0983638406 |
- |
- |
PREDICTED: cytochrome P450 78A9 [Jatropha curcas] |
| 5 |
Hb_000840_230 |
0.1134019585 |
- |
- |
RING-H2 finger protein ATL1L, putative [Ricinus communis] |
| 6 |
Hb_000318_320 |
0.1186610592 |
- |
- |
PREDICTED: transmembrane protein 45B [Jatropha curcas] |
| 7 |
Hb_107495_050 |
0.1316526266 |
transcription factor |
TF Family: MIKC |
mads box protein, putative [Ricinus communis] |
| 8 |
Hb_170979_010 |
0.1367328986 |
- |
- |
PREDICTED: peroxidase 4-like [Jatropha curcas] |
| 9 |
Hb_002078_210 |
0.1414247771 |
- |
- |
PREDICTED: cytochrome P450 78A5-like [Jatropha curcas] |
| 10 |
Hb_009393_010 |
0.1430844073 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF017 [Jatropha curcas] |
| 11 |
Hb_077562_010 |
0.1456459493 |
- |
- |
PREDICTED: WAT1-related protein At5g07050 [Jatropha curcas] |
| 12 |
Hb_013862_010 |
0.150841657 |
- |
- |
PREDICTED: uncharacterized protein LOC105650642 [Jatropha curcas] |
| 13 |
Hb_009230_050 |
0.1523293835 |
- |
- |
PREDICTED: transcription factor bHLH113-like [Jatropha curcas] |
| 14 |
Hb_008296_010 |
0.1524651695 |
- |
- |
- |
| 15 |
Hb_001784_010 |
0.152535278 |
- |
- |
nutrient reservoir, putative [Ricinus communis] |
| 16 |
Hb_010615_010 |
0.1529183109 |
- |
- |
PREDICTED: reticuline oxidase-like protein [Jatropha curcas] |
| 17 |
Hb_000103_280 |
0.1534709042 |
transcription factor |
TF Family: HSF |
Heat shock factor protein HSF30, putative [Ricinus communis] |
| 18 |
Hb_070850_010 |
0.1536878258 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor 1B-like [Jatropha curcas] |
| 19 |
Hb_007556_010 |
0.1538261395 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360-like isoform X2 [Populus euphratica] |
| 20 |
Hb_003938_150 |
0.1540597256 |
- |
- |
hypothetical protein JCGZ_17711 [Jatropha curcas] |