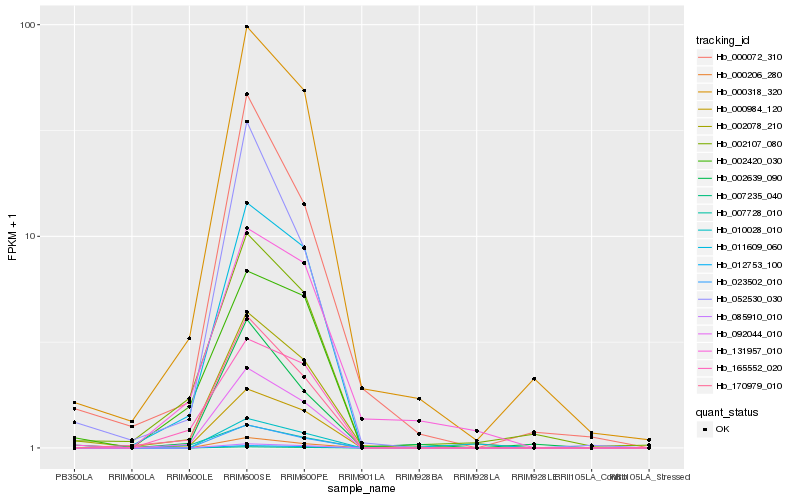
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002078_210 |
0.0 |
- |
- |
PREDICTED: cytochrome P450 78A5-like [Jatropha curcas] |
| 2 |
Hb_000318_320 |
0.1130639192 |
- |
- |
PREDICTED: transmembrane protein 45B [Jatropha curcas] |
| 3 |
Hb_011609_060 |
0.119291106 |
- |
- |
PREDICTED: cytokinin hydroxylase [Jatropha curcas] |
| 4 |
Hb_092044_010 |
0.1267239814 |
- |
- |
PREDICTED: uncharacterized protein LOC105636336 [Jatropha curcas] |
| 5 |
Hb_170979_010 |
0.1327338021 |
- |
- |
PREDICTED: peroxidase 4-like [Jatropha curcas] |
| 6 |
Hb_002107_080 |
0.133108357 |
- |
- |
JHL20J20.3 [Jatropha curcas] |
| 7 |
Hb_000984_120 |
0.1398404042 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 8 |
Hb_000072_310 |
0.1414247771 |
transcription factor |
TF Family: bZIP |
PREDICTED: basic leucine zipper 43-like [Jatropha curcas] |
| 9 |
Hb_002420_030 |
0.1479016575 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 10 |
Hb_052530_030 |
0.1565745514 |
- |
- |
PREDICTED: cytochrome P450 78A9 [Jatropha curcas] |
| 11 |
Hb_010028_010 |
0.1595700391 |
- |
- |
serine/threonine-protein kinase bri1, putative [Ricinus communis] |
| 12 |
Hb_007235_040 |
0.1596503268 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 13 |
Hb_131957_010 |
0.1605753442 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 14 |
Hb_085910_010 |
0.1621747128 |
- |
- |
PREDICTED: uncharacterized protein LOC104106381 [Nicotiana tomentosiformis] |
| 15 |
Hb_002639_090 |
0.1624562917 |
- |
- |
hypothetical protein JCGZ_08134 [Jatropha curcas] |
| 16 |
Hb_000206_280 |
0.1628742963 |
- |
- |
Uncharacterized protein TCM_031897 [Theobroma cacao] |
| 17 |
Hb_023502_010 |
0.1634376402 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 18 |
Hb_165552_020 |
0.1635494129 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 19 |
Hb_012753_100 |
0.1646744263 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_007728_010 |
0.1668867982 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Jatropha curcas] |