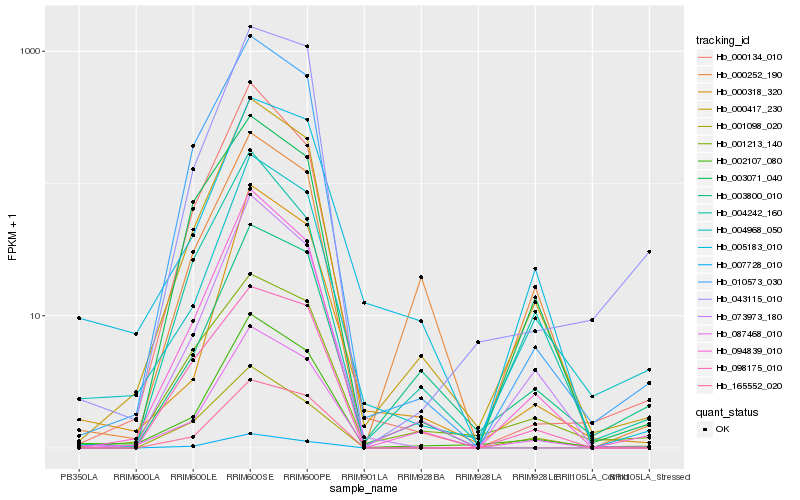
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000417_230 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_094839_010 |
0.0597564172 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002107_080 |
0.0730938691 |
- |
- |
JHL20J20.3 [Jatropha curcas] |
| 4 |
Hb_073973_180 |
0.0800821313 |
- |
- |
PREDICTED: WAT1-related protein At4g08290 isoform X1 [Populus euphratica] |
| 5 |
Hb_010573_030 |
0.0931953011 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_003071_040 |
0.1119775414 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF017 [Jatropha curcas] |
| 7 |
Hb_004968_050 |
0.1133993157 |
- |
- |
PREDICTED: putative lipid-transfer protein DIR1 [Jatropha curcas] |
| 8 |
Hb_007728_010 |
0.120168855 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Jatropha curcas] |
| 9 |
Hb_001213_140 |
0.1206371276 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 10 |
Hb_000134_010 |
0.122991818 |
- |
- |
- |
| 11 |
Hb_004242_160 |
0.1248870711 |
transcription factor |
TF Family: GRAS |
PREDICTED: chitin-inducible gibberellin-responsive protein 1-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_000318_320 |
0.1258517679 |
- |
- |
PREDICTED: transmembrane protein 45B [Jatropha curcas] |
| 13 |
Hb_165552_020 |
0.1289566927 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 14 |
Hb_003800_010 |
0.1296532062 |
- |
- |
PREDICTED: cysteine-rich and transmembrane domain-containing protein A-like [Jatropha curcas] |
| 15 |
Hb_087468_010 |
0.1301047214 |
- |
- |
PREDICTED: uncharacterized protein LOC100246093 [Vitis vinifera] |
| 16 |
Hb_098175_010 |
0.1301283395 |
- |
- |
hypothetical protein JCGZ_23771 [Jatropha curcas] |
| 17 |
Hb_005183_010 |
0.1308774959 |
- |
- |
PREDICTED: probable calcium-binding protein CML17 [Malus domestica] |
| 18 |
Hb_001098_020 |
0.1314228173 |
- |
- |
PREDICTED: transcription factor bHLH123-like isoform X1 [Populus euphratica] |
| 19 |
Hb_000252_190 |
0.1319206872 |
- |
- |
PREDICTED: uncharacterized protein LOC105644975 [Jatropha curcas] |
| 20 |
Hb_043115_010 |
0.1322609703 |
- |
- |
hypothetical protein JCGZ_16710 [Jatropha curcas] |