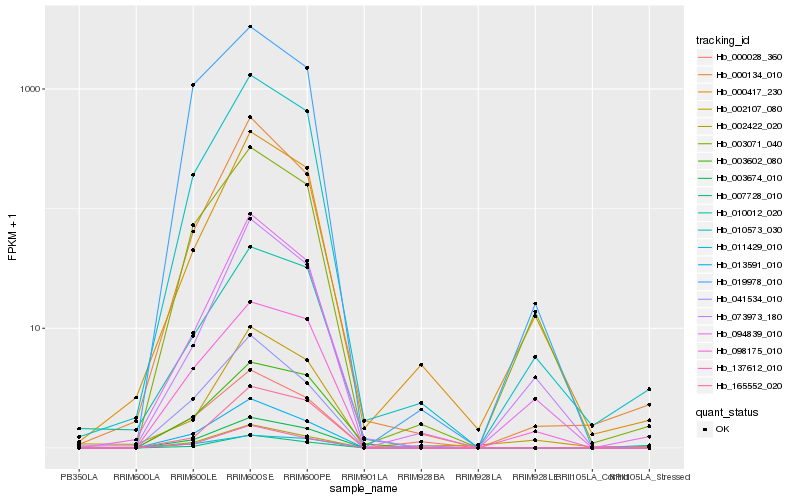
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010573_030 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_007728_010 |
0.0645886986 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Jatropha curcas] |
| 3 |
Hb_002422_020 |
0.0675245216 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL5 [Jatropha curcas] |
| 4 |
Hb_137612_010 |
0.0733028361 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 5 |
Hb_013591_010 |
0.0737378955 |
- |
- |
Mitogen-activated protein kinase kinase kinase 1 [Glycine soja] |
| 6 |
Hb_000134_010 |
0.0808724129 |
- |
- |
- |
| 7 |
Hb_094839_010 |
0.085948763 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_165552_020 |
0.0929326011 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 9 |
Hb_000417_230 |
0.0931953011 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_010012_020 |
0.0958821356 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_098175_010 |
0.1011964186 |
- |
- |
hypothetical protein JCGZ_23771 [Jatropha curcas] |
| 12 |
Hb_003674_010 |
0.1013865324 |
- |
- |
purine transporter, putative [Ricinus communis] |
| 13 |
Hb_003071_040 |
0.1017827474 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF017 [Jatropha curcas] |
| 14 |
Hb_003602_080 |
0.1092499995 |
- |
- |
hypothetical protein POPTR_0002s10890g [Populus trichocarpa] |
| 15 |
Hb_041534_010 |
0.1133531733 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF017 [Jatropha curcas] |
| 16 |
Hb_011429_010 |
0.1166035986 |
- |
- |
hypothetical protein POPTR_0188s00230g [Populus trichocarpa] |
| 17 |
Hb_002107_080 |
0.116667932 |
- |
- |
JHL20J20.3 [Jatropha curcas] |
| 18 |
Hb_073973_180 |
0.11729845 |
- |
- |
PREDICTED: WAT1-related protein At4g08290 isoform X1 [Populus euphratica] |
| 19 |
Hb_019978_010 |
0.123605565 |
- |
- |
PREDICTED: glycerophosphodiester phosphodiesterase protein kinase domain-containing GDPDL2-like isoform X2 [Jatropha curcas] |
| 20 |
Hb_000028_360 |
0.1237120982 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |