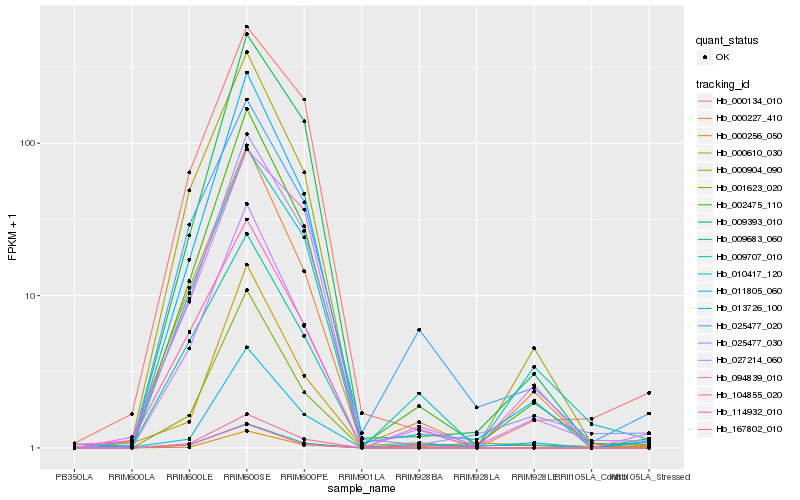
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_025477_030 |
0.0 |
transcription factor |
TF Family: ERF |
dehydration-responsive element-binding protein 2 [Manihot esculenta] |
| 2 |
Hb_009393_010 |
0.070163848 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF017 [Jatropha curcas] |
| 3 |
Hb_002475_110 |
0.0723128501 |
- |
- |
E3 ubiquitin ligase PUB14, putative [Ricinus communis] |
| 4 |
Hb_013726_100 |
0.0743796047 |
- |
- |
PREDICTED: BON1-associated protein 2 [Jatropha curcas] |
| 5 |
Hb_167802_010 |
0.0805262008 |
- |
- |
PREDICTED: receptor-like protein 12 [Populus euphratica] |
| 6 |
Hb_000134_010 |
0.0850363761 |
- |
- |
- |
| 7 |
Hb_000904_090 |
0.0945224072 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF109-like [Jatropha curcas] |
| 8 |
Hb_009683_060 |
0.0966707735 |
- |
- |
- |
| 9 |
Hb_000256_050 |
0.1013018214 |
- |
- |
chitinase, putative [Ricinus communis] |
| 10 |
Hb_010417_120 |
0.1053633522 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000227_410 |
0.1059518714 |
- |
- |
PREDICTED: uncharacterized protein LOC105631970 [Jatropha curcas] |
| 12 |
Hb_027214_060 |
0.1059613901 |
- |
- |
PREDICTED: exocyst complex component EXO70A1-like [Jatropha curcas] |
| 13 |
Hb_001623_020 |
0.1062022075 |
- |
- |
9-cis-epoxycarotenoid dioxygenase, putative [Ricinus communis] |
| 14 |
Hb_000610_030 |
0.1098905651 |
- |
- |
hypothetical protein MIMGU_mgv1a0038491mg, partial [Erythranthe guttata] |
| 15 |
Hb_009707_010 |
0.1161026375 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: exocyst complex component EXO70B1-like [Jatropha curcas] |
| 16 |
Hb_094839_010 |
0.1165320155 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_104855_020 |
0.1212418995 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 18 |
Hb_025477_020 |
0.1238046679 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF027 [Jatropha curcas] |
| 19 |
Hb_114932_010 |
0.124772172 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 20 |
Hb_011805_060 |
0.1249182234 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |