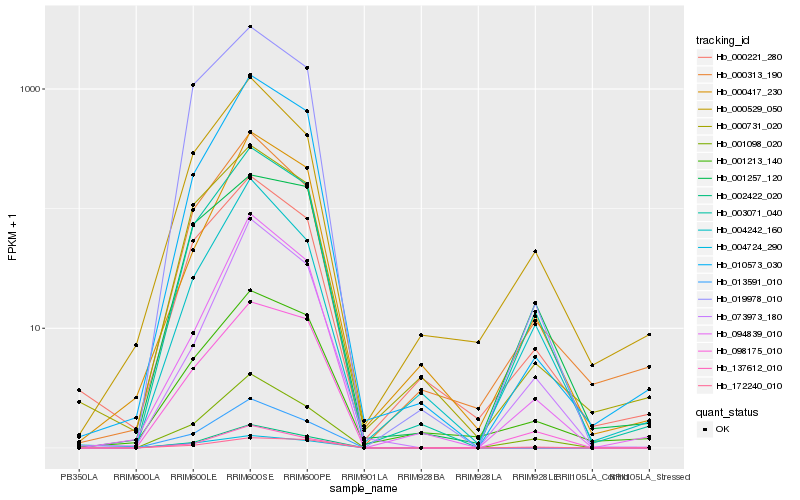
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003071_040 |
0.0 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF017 [Jatropha curcas] |
| 2 |
Hb_001098_020 |
0.0673500839 |
- |
- |
PREDICTED: transcription factor bHLH123-like isoform X1 [Populus euphratica] |
| 3 |
Hb_098175_010 |
0.0848216773 |
- |
- |
hypothetical protein JCGZ_23771 [Jatropha curcas] |
| 4 |
Hb_000313_190 |
0.0908066939 |
- |
- |
PREDICTED: uncharacterized protein LOC105636896 [Jatropha curcas] |
| 5 |
Hb_000731_020 |
0.0988263822 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein ZAT10-like [Jatropha curcas] |
| 6 |
Hb_010573_030 |
0.1017827474 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000529_050 |
0.1023698648 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 5 [Jatropha curcas] |
| 8 |
Hb_000221_280 |
0.1026472451 |
- |
- |
PREDICTED: protein EXORDIUM-like [Populus euphratica] |
| 9 |
Hb_019978_010 |
0.1075757175 |
- |
- |
PREDICTED: glycerophosphodiester phosphodiesterase protein kinase domain-containing GDPDL2-like isoform X2 [Jatropha curcas] |
| 10 |
Hb_172240_010 |
0.1078044415 |
- |
- |
PREDICTED: uncharacterized protein LOC105795660 [Gossypium raimondii] |
| 11 |
Hb_004242_160 |
0.1118590061 |
transcription factor |
TF Family: GRAS |
PREDICTED: chitin-inducible gibberellin-responsive protein 1-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_000417_230 |
0.1119775414 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001213_140 |
0.1124640313 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 14 |
Hb_002422_020 |
0.1140389101 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL5 [Jatropha curcas] |
| 15 |
Hb_013591_010 |
0.1145632893 |
- |
- |
Mitogen-activated protein kinase kinase kinase 1 [Glycine soja] |
| 16 |
Hb_001257_120 |
0.115340499 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 9 [Jatropha curcas] |
| 17 |
Hb_094839_010 |
0.1164577292 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_137612_010 |
0.1269389735 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 19 |
Hb_073973_180 |
0.1275000521 |
- |
- |
PREDICTED: WAT1-related protein At4g08290 isoform X1 [Populus euphratica] |
| 20 |
Hb_004724_290 |
0.1327264762 |
transcription factor |
TF Family: NAC |
- |