| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
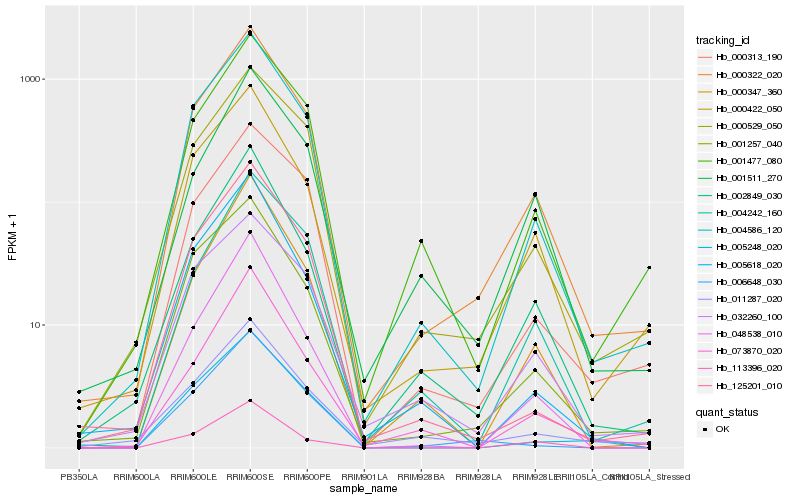
Hb_000322_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105637914 [Jatropha curcas] |
| 2 |
Hb_004586_120 |
0.0503280083 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 5 [Jatropha curcas] |
| 3 |
Hb_000422_050 |
0.0680289809 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 9 [Jatropha curcas] |
| 4 |
Hb_002849_030 |
0.0787909784 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0014s09190g [Populus trichocarpa] |
| 5 |
Hb_001257_040 |
0.084482719 |
- |
- |
PREDICTED: uncharacterized protein LOC105640055 [Jatropha curcas] |
| 6 |
Hb_000347_360 |
0.0886949271 |
- |
- |
PREDICTED: linoleate 13S-lipoxygenase 3-1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000529_050 |
0.0917771064 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 5 [Jatropha curcas] |
| 8 |
Hb_001477_080 |
0.0933733614 |
- |
- |
hypothetical protein POPTR_0017s04860g [Populus trichocarpa] |
| 9 |
Hb_048538_010 |
0.1016592438 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Prunus mume] |
| 10 |
Hb_073870_020 |
0.1027224182 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000, partial [Jatropha curcas] |
| 11 |
Hb_125201_010 |
0.1064939151 |
- |
- |
hypothetical protein POPTR_0004s03690g [Populus trichocarpa] |
| 12 |
Hb_000313_190 |
0.1079508308 |
- |
- |
PREDICTED: uncharacterized protein LOC105636896 [Jatropha curcas] |
| 13 |
Hb_004242_160 |
0.1106042945 |
transcription factor |
TF Family: GRAS |
PREDICTED: chitin-inducible gibberellin-responsive protein 1-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_005618_020 |
0.1115431529 |
- |
- |
hypothetical protein RCOM_1342750 [Ricinus communis] |
| 15 |
Hb_011287_020 |
0.1127688149 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 16 |
Hb_032260_100 |
0.1133101096 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 53 [Jatropha curcas] |
| 17 |
Hb_006648_030 |
0.1149137613 |
- |
- |
Indole-3-acetic acid-amido synthetase GH3.3, putative [Ricinus communis] |
| 18 |
Hb_005248_020 |
0.1156070703 |
- |
- |
hypothetical protein JCGZ_23770 [Jatropha curcas] |
| 19 |
Hb_001511_270 |
0.1181198034 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 29 [Jatropha curcas] |
| 20 |
Hb_113396_020 |
0.118208494 |
- |
- |
Mitogen-activated protein kinase kinase kinase 1 [Glycine soja] |