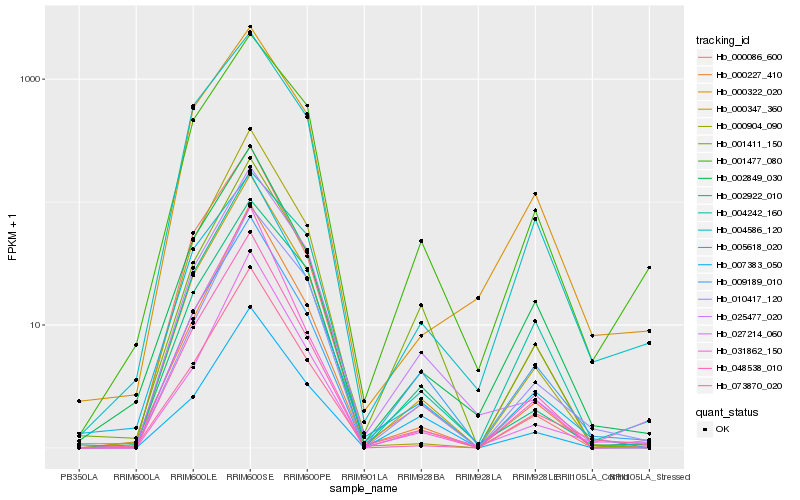
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000347_360 |
0.0 |
- |
- |
PREDICTED: linoleate 13S-lipoxygenase 3-1, chloroplastic [Jatropha curcas] |
| 2 |
Hb_073870_020 |
0.0611048067 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000, partial [Jatropha curcas] |
| 3 |
Hb_048538_010 |
0.0623933827 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Prunus mume] |
| 4 |
Hb_000227_410 |
0.0696443169 |
- |
- |
PREDICTED: uncharacterized protein LOC105631970 [Jatropha curcas] |
| 5 |
Hb_002849_030 |
0.0733870117 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0014s09190g [Populus trichocarpa] |
| 6 |
Hb_000322_020 |
0.0886949271 |
- |
- |
PREDICTED: uncharacterized protein LOC105637914 [Jatropha curcas] |
| 7 |
Hb_027214_060 |
0.0899367113 |
- |
- |
PREDICTED: exocyst complex component EXO70A1-like [Jatropha curcas] |
| 8 |
Hb_009189_010 |
0.0904571084 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 33 [Jatropha curcas] |
| 9 |
Hb_000904_090 |
0.0905148718 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF109-like [Jatropha curcas] |
| 10 |
Hb_031862_150 |
0.0926949969 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 5 [Jatropha curcas] |
| 11 |
Hb_004586_120 |
0.0930555284 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 5 [Jatropha curcas] |
| 12 |
Hb_010417_120 |
0.0983646313 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_004242_160 |
0.1026273208 |
transcription factor |
TF Family: GRAS |
PREDICTED: chitin-inducible gibberellin-responsive protein 1-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_025477_020 |
0.1069289058 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF027 [Jatropha curcas] |
| 15 |
Hb_001477_080 |
0.1075616126 |
- |
- |
hypothetical protein POPTR_0017s04860g [Populus trichocarpa] |
| 16 |
Hb_002922_010 |
0.1083263669 |
- |
- |
heavy-metal-associated domain-containing family protein [Populus trichocarpa] |
| 17 |
Hb_005618_020 |
0.1092442694 |
- |
- |
hypothetical protein RCOM_1342750 [Ricinus communis] |
| 18 |
Hb_001411_150 |
0.1110728451 |
- |
- |
Dormancy/auxin associated family protein, putative [Theobroma cacao] |
| 19 |
Hb_000086_600 |
0.1115663515 |
- |
- |
- |
| 20 |
Hb_007383_050 |
0.1155729333 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF061 [Jatropha curcas] |